Identification of ancient remains through genomic sequencing
- PMID: 18426903
- PMCID: PMC2493433
- DOI: 10.1101/gr.076091.108
Identification of ancient remains through genomic sequencing
Erratum in
- Genome Res. 2008 Nov;18(11):1859
Abstract
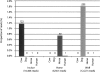
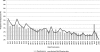
Studies of ancient DNA have been hindered by the preciousness of remains, the small quantities of undamaged DNA accessible, and the limitations associated with conventional PCR amplification. In these studies, we developed and applied a genomewide adapter-mediated emulsion PCR amplification protocol for ancient mammalian samples estimated to be between 45,000 and 69,000 yr old. Using 454 Life Sciences (Roche) and Illumina sequencing (formerly Solexa sequencing) technologies, we examined over 100 megabases of DNA from amplified extracts, revealing unbiased sequence coverage with substantial amounts of nonredundant nuclear sequences from the sample sources and negligible levels of human contamination. We consistently recorded over 500-fold increases, such that nanogram quantities of starting material could be amplified to microgram quantities. Application of our protocol to a 50,000-yr-old uncharacterized bone sample that was unsuccessful in mitochondrial PCR provided sufficient nuclear sequences for comparison with extant mammals and subsequent phylogenetic classification of the remains. The combined use of emulsion PCR amplification and high-throughput sequencing allows for the generation of large quantities of DNA sequence data from ancient remains. Using such techniques, even small amounts of ancient remains with low levels of endogenous DNA preservation may yield substantial quantities of nuclear DNA, enabling novel applications of ancient DNA genomics to the investigation of extinct phyla.
Figures



References
-
- Briggs A.W., Stenzel U., Johnson P.L., Green R.E., Kelso J., Prufer K., Meyer M., Krause J., Ronan M.T., Lachmann M., Stenzel U., Johnson P.L., Green R.E., Kelso J., Prufer K., Meyer M., Krause J., Ronan M.T., Lachmann M., Johnson P.L., Green R.E., Kelso J., Prufer K., Meyer M., Krause J., Ronan M.T., Lachmann M., Green R.E., Kelso J., Prufer K., Meyer M., Krause J., Ronan M.T., Lachmann M., Kelso J., Prufer K., Meyer M., Krause J., Ronan M.T., Lachmann M., Prufer K., Meyer M., Krause J., Ronan M.T., Lachmann M., Meyer M., Krause J., Ronan M.T., Lachmann M., Krause J., Ronan M.T., Lachmann M., Ronan M.T., Lachmann M., Lachmann M., et al. Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl. Acad. Sci. 2007;104:14616–14621. - PMC - PubMed
-
- Derevianko A.P., Shunkov M.V., Agadjaniav A.K., Baryshnikov G.F., Malaeva E.M., Ulianov V.A., Kulik N.A., Postnov A.V., Anoikin A.A., Shunkov M.V., Agadjaniav A.K., Baryshnikov G.F., Malaeva E.M., Ulianov V.A., Kulik N.A., Postnov A.V., Anoikin A.A., Agadjaniav A.K., Baryshnikov G.F., Malaeva E.M., Ulianov V.A., Kulik N.A., Postnov A.V., Anoikin A.A., Baryshnikov G.F., Malaeva E.M., Ulianov V.A., Kulik N.A., Postnov A.V., Anoikin A.A., Malaeva E.M., Ulianov V.A., Kulik N.A., Postnov A.V., Anoikin A.A., Ulianov V.A., Kulik N.A., Postnov A.V., Anoikin A.A., Kulik N.A., Postnov A.V., Anoikin A.A., Postnov A.V., Anoikin A.A., Anoikin A.A. Paleoenvironment and paleolithic human occupation of Gorny Altai (Subsistence and adaptation in the vicinity of Denisova Cave) Russian Academy of Sciences; Novosibirsk: 2003.
-
- Ewing B., Green P., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. - PubMed
-
- Ewing B., Hillier L., Wendl M.C., Green P., Hillier L., Wendl M.C., Green P., Wendl M.C., Green P., Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
