Transmission of HIV-1 Gag immune escape mutations is associated with reduced viral load in linked recipients
- PMID: 18426987
- PMCID: PMC2373834
- DOI: 10.1084/jem.20072457
Transmission of HIV-1 Gag immune escape mutations is associated with reduced viral load in linked recipients
Abstract
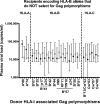
In a study of 114 epidemiologically linked Zambian transmission pairs, we evaluated the impact of human leukocyte antigen class I (HLA-I)-associated amino acid polymorphisms, presumed to reflect cytotoxic T lymphocyte (CTL) escape in Gag and Nef of the virus transmitted from the chronically infected donor, on the plasma viral load (VL) in matched recipients 6 mo after infection. CTL escape mutations in Gag and Nef were seen in the donors, which were subsequently transmitted to recipients, largely unchanged soon after infection. We observed a significant correlation between the number of Gag escape mutations targeted by specific HLA-B allele-restricted CTLs and reduced VLs in the recipients. This negative correlation was most evident in newly infected individuals, whose HLA alleles were unable to effectively target Gag and select for CTL escape mutations in this gene. Nef mutations in the donor had no impact on VL in the recipient. Thus, broad Gag-specific CTL responses capable of driving virus escape in the donor may be of clinical benefit to both the donor and recipient. In addition to their direct implications for HIV-1 vaccine design, these data suggest that CTL-induced viral polymorphisms and their associated in vivo viral fitness costs could have a significant impact on HIV-1 pathogenesis.
Figures



References
-
- Goulder, P.J., and D.I. Watkins. 2004. HIV and SIV CTL escape: implications for vaccine design. Nat. Rev. Immunol. 4:630–640. - PubMed
-
- Goulder, P.J., R.E. Phillips, R.A. Colbert, S. McAdam, G. Ogg, M.A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, et al. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212–217. - PubMed
-
- Ramduth, D., P. Chetty, N.C. Mngquandaniso, N. Nene, J.D. Harlow, I. Honeyborne, N. Ntumba, S. Gappoo, C. Henry, P. Jeena, et al. 2005. Differential immunogenicity of HIV-1 clade C proteins in eliciting CD8+ and CD4+ cell responses. J. Infect. Dis. 192:1588–1596. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

