Global transcriptomic analysis of Cyanothece 51142 reveals robust diurnal oscillation of central metabolic processes
- PMID: 18427117
- PMCID: PMC2329701
- DOI: 10.1073/pnas.0711068105
Global transcriptomic analysis of Cyanothece 51142 reveals robust diurnal oscillation of central metabolic processes
Abstract
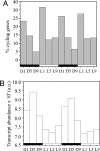
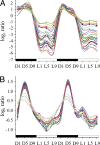
Cyanobacteria are photosynthetic organisms and are the only prokaryotes known to have a circadian lifestyle. Unicellular diazotrophic cyanobacteria such as Cyanothece sp. ATCC 51142 produce oxygen and can also fix atmospheric nitrogen, a process exquisitely sensitive to oxygen. To accommodate such antagonistic processes, the intracellular environment of Cyanothece oscillates between aerobic and anaerobic conditions during a day-night cycle. This is accomplished by temporal separation of the two processes: photosynthesis during the day and nitrogen fixation at night. Although previous studies have examined periodic changes in transcript levels for a limited number of genes in Cyanothece and other unicellular diazotrophic cyanobacteria, a comprehensive study of transcriptional activity in a nitrogen-fixing cyanobacterium is necessary to understand the impact of the temporal separation of photosynthesis and nitrogen fixation on global gene regulation and cellular metabolism. We have examined the expression patterns of nearly 5,000 genes in Cyanothece 51142 during two consecutive diurnal periods. Our analysis showed that approximately 30% of these genes exhibited robust oscillating expression profiles. Interestingly, this set included genes for almost all central metabolic processes in Cyanothece 51142. A transcriptional network of all genes with significantly oscillating transcript levels revealed that the majority of genes encoding enzymes in numerous individual biochemical pathways, such as glycolysis, oxidative pentose phosphate pathway, and glycogen metabolism, were coregulated and maximally expressed at distinct phases during the diurnal cycle. These studies provide a comprehensive picture of how a physiologically relevant diurnal light-dark cycle influences the metabolism in a photosynthetic bacterium.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Herzog ED. Neurons and networks in daily rhythms. Nat Rev Neurosci. 2007;8:790–802. - PubMed
-
- Hotta CT, et al. Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ. 2007;30:333–349. - PubMed
-
- Dunlap JC, Loros JJ. How fungi keep time: circadian system in Neurospora and other fungi. Curr Opin Microbiol. 2006;9:579–587. - PubMed
-
- Morse DS, Fritz L, Hastings JW. What is the clock? Translational regulation of circadian bioluminescence. Trends Biochem Sci. 1990;15:262–265. - PubMed
-
- Johnson CH, Golden SS, Ishiura M, Kondo T. Circadian clocks in prokaryotes. Mol Microbiol. 1996;21:5–11. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

