Lipid spirals in Bacillus subtilis and their role in cell division
- PMID: 18430139
- PMCID: PMC2408660
- DOI: 10.1111/j.1365-2958.2008.06236.x
Lipid spirals in Bacillus subtilis and their role in cell division
Abstract
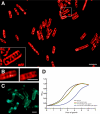
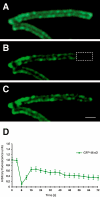
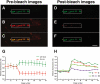
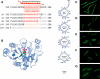
The fluid mosaic model of membrane structure has been revised in recent years as it has become evident that domains of different lipid composition are present in eukaryotic and prokaryotic cells. Using membrane binding fluorescent dyes, we demonstrate the presence of lipid spirals extending along the long axis of cells of the rod-shaped bacterium Bacillus subtilis. These spiral structures are absent from cells in which the synthesis of phosphatidylglycerol is disrupted, suggesting an enrichment in anionic phospholipids. Green fluorescent protein fusions of the cell division protein MinD also form spiral structures and these were shown by fluorescence resonance energy transfer to be coincident with the lipid spirals. These data indicate a higher level of membrane lipid organization than previously observed and a primary role for lipid spirals in determining the site of cell division in bacterial cells.
Figures


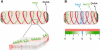
References
-
- Barák I, Wilkinson AJ. Division site recognition in Escherichia coli and Bacillus subtilis. FEMS Microbiol Rev. 2007;31:311–326. - PubMed
-
- Ben-Yehuda S, Losick R. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell. 2002;109:257–266. - PubMed
-
- Brumback AC, Lieber JL, Angleson JK, Betz WJ. Using FM1-43 to study neuropeptide granule dynamics and exocytosis. Methods. 2004;33:287–294. - PubMed
-
- Campo N, Tjalsma H, Buist G, Stepniak D, Meijer M, Veenhuis M, et al. Subcellular sites for bacterial protein export. Mol Microbiol. 2004;53:1583–1599. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

