Widespread duplications in the genomes of laboratory stocks of Dictyostelium discoideum
- PMID: 18430225
- PMCID: PMC2643946
- DOI: 10.1186/gb-2008-9-4-r75
Widespread duplications in the genomes of laboratory stocks of Dictyostelium discoideum
Abstract
Background: Duplications of stretches of the genome are an important source of individual genetic variation, but their unrecognized presence in laboratory organisms would be a confounding variable for genetic analysis.
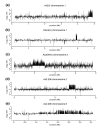
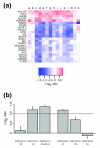
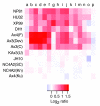
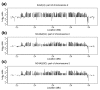
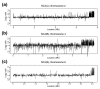
Results: We report here that duplications of 15 kb or more are common in the genome of the social amoeba Dictyostelium discoideum. Most stocks of the axenic 'workhorse' strains Ax2 and Ax3/4 obtained from different laboratories can be expected to carry different duplications. The auxotrophic strains DH1 and JH10 also bear previously unreported duplications. Strain Ax3/4 is known to carry a large duplication on chromosome 2 and this structure shows evidence of continuing instability; we find a further variable duplication on chromosome 5. These duplications are lacking in Ax2, which has instead a small duplication on chromosome 1. Stocks of the type isolate NC4 are similarly variable, though we have identified some approximating the assumed ancestral genotype. More recent wild-type isolates are almost without large duplications, but we can identify small deletions or regions of high divergence, possibly reflecting responses to local selective pressures. Duplications are scattered through most of the genome, and can be stable enough to reconstruct genealogies spanning decades of the history of the NC4 lineage. The expression level of many duplicated genes is increased with dosage, but for others it appears that some form of dosage compensation occurs.
Conclusion: The genetic variation described here must underlie some of the phenotypic variation observed between strains from different laboratories. We suggest courses of action to alleviate the problem.
Figures




References
-
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. - DOI - PubMed
-
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez, Gratacas, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F. et al.Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. - DOI - PMC - PubMed
-
- Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson MR, Muenter M, Baptista M, Miller D, Blancato J, Hardy J, Gwinn-Hardy K. alpha-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

