Paramutation: epigenetic instructions passed across generations
- PMID: 18430919
- PMCID: PMC2323780
- DOI: 10.1093/genetics/178.4.1839
Paramutation: epigenetic instructions passed across generations
Figures
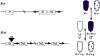

References
-
- Alleman, M., and J. Doctor, 2000. Genomic imprinting in plants: observations and evolutionary implications. Plant Mol. Biol. 43 147–161. - PubMed
-
- Alleman, M., L. Sidorenko, K. McGinnis, V. Seshadri, J. E. Dorweiler et al., 2006. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 442 295–298. - PubMed
-
- Brink, R. A., 1973. Paramutation. Annu. Rev. Genet. 7 129–152. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

