Genetic analysis of the Caenorhabditis elegans GLH family of P-granule proteins
- PMID: 18430929
- PMCID: PMC2323790
- DOI: 10.1534/genetics.107.083469
Genetic analysis of the Caenorhabditis elegans GLH family of P-granule proteins
Abstract
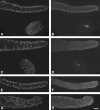
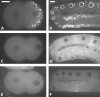
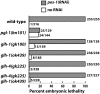
The Vasa DEAD-box helicases are widespread markers of germ cells across species, and in some organisms have been shown to be essential for germ-cell formation and development. In contrast to the single Vasa gene in most systems analyzed, Caenorhabditis elegans has four Vasa family members, the germline helicases GLH-1, GLH-2, GLH-3, and GLH-4. Our analysis of deletion alleles of each glh gene demonstrates that GLH-1 is the key member of the family: loss of GLH-1 function causes sterility that is mainly maternal effect, is manifested predominantly at elevated temperature, and is due to reduced germ-cell proliferation and impaired formation of both sperm and oocytes. The other GLHs are not essential. However, GLH-4 serves redundant roles with GLH-1: loss of both genes' function causes glh-1-like sterility at all temperatures. Molecular epistasis analysis demonstrates that GLH-1 and GLH-4 are required for proper association of the PGL family of proteins with P granules, suggesting a pathway of P-granule assembly in which the GLHs are upstream of the PGL proteins and the mRNA cap-binding protein IFE-1. While loss of some P-granule components causes worms to be defective in RNA interference, loss of GLH-1 and GLH-4 does not compromise RNAi. Thus, RNAi likely does not require intact P granules but instead relies on particular P-granule factors. We discuss the evolution of the Vasa/GLH genes and current views of their functions and the assembly and roles of germ granules among species.
Figures


References
-
- Breitwieser, W., F. H. Markussen, H. Horstmann and A. Ephrussi, 1996. Oskar protein interaction with Vasa represents an essential step in polar granule assembly. Genes Dev. 10 2179–2188. - PubMed
-
- Carmichael, J. B., C. Stoica, H. Parker, J. M. McCaffery, A. J. Simmonds et al., 2006. RNA interference effector proteins localize to mobile cytoplasmic puncta in Schizosaccharomyces pombe. Traffic 7 1032–1044. - PubMed
-
- Chan, S. P., and F. J. Slack, 2006. MicroRNA-mediated silencing inside P-bodies. RNA Biol. 3 97–100. - PubMed
-
- Cheeks, R. J., J. C. Canman, W. N. Gabriel, N. Meyer, S. Strome et al., 2004. C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes. Curr. Biol. 14 851–862. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

