Raw mediates antagonism of AP-1 activity in Drosophila
- PMID: 18430930
- PMCID: PMC2323791
- DOI: 10.1534/genetics.107.086298
Raw mediates antagonism of AP-1 activity in Drosophila
Abstract
High baselines of transcription factor activities represent fundamental obstacles to regulated signaling. Here we show that in Drosophila, quenching of basal activator protein 1 (AP-1) transcription factor activity serves as a prerequisite to its tight spatial and temporal control by the JNK (Jun N-terminal kinase) signaling cascade. Our studies indicate that the novel raw gene product is required to limit AP-1 activity to leading edge epidermal cells during embryonic dorsal closure. In addition, we provide the first evidence that the epidermis has a Basket JNK-independent capacity to activate AP-1 targets and that raw function is required broadly throughout the epidermis to antagonize this activity. Finally, our mechanistic studies of the three dorsal-open group genes [raw, ribbon (rib), and puckered (puc)] indicate that these gene products provide at least two tiers of JNK/AP-1 regulation. In addition to Puckered phosphatase function in leading edge epidermal cells as a negative-feedback regulator of JNK signaling, the three dorsal-open group gene products (Raw, Ribbon, and Puckered) are required more broadly in the dorsolateral epidermis to quench a basal, signaling-independent activity of the AP-1 transcription factor.
Figures

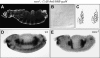

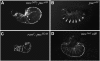


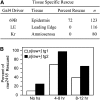
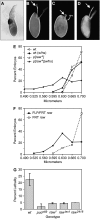

References
-
- Arora, K., H. Dai, S. Kazuko, J. Jamal, M. O'Connor et al., 1995. The Drosophila schnurri gene acts in the Dpp/TGF beta signaling pathway and encodes a transcription factor homologous to the human MBP family. Cell 81 781–790. - PubMed
-
- Blake, K. J., G. Myette and J. Jack, 1998. The products of ribbon and raw are necessary for proper cell shape and cellular localization of nonmuscle myosin in Drosophila. Dev. Biol. 203 177–188. - PubMed
-
- Blake, K. J., G. Myette and J. Jack, 1999. ribbon, raw, and zipper have distinct functions in reshaping the Drosophila cytoskeleton. Dev. Genes Evol. 209 555–559. - PubMed
-
- Bradley, P. L., and D. J. Andrew, 2001. ribbon encodes a novel BTB/POZ protein required for directed cell migration in Drosophila melanogaster. Development 128 3001–3015. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

