Inferring microRNA activities by combining gene expression with microRNA target prediction
- PMID: 18431476
- PMCID: PMC2291556
- DOI: 10.1371/journal.pone.0001989
Inferring microRNA activities by combining gene expression with microRNA target prediction
Abstract
Background: MicroRNAs (miRNAs) play crucial roles in a variety of biological processes via regulating expression of their target genes at the mRNA level. A number of computational approaches regarding miRNAs have been proposed, but most of them focus on miRNA gene finding or target predictions. Little computational work has been done to investigate the effective regulation of miRNAs.
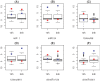
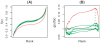
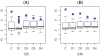
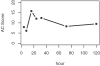
Methodology/principal findings: We propose a method to infer the effective regulatory activities of miRNAs by integrating microarray expression data with miRNA target predictions. The method is based on the idea that regulatory activity changes of miRNAs could be reflected by the expression changes of their target transcripts measured by microarray. To validate this method, we apply it to the microarray data sets that measure gene expression changes in cell lines after transfection or inhibition of several specific miRNAs. The results indicate that our method can detect activity enhancement of the transfected miRNAs as well as activity reduction of the inhibited miRNAs with high sensitivity and specificity. Furthermore, we show that our inference is robust with respect to false positives of target prediction.
Conclusions/significance: A huge amount of gene expression data sets are available in the literature, but miRNA regulation underlying these data sets is largely unknown. The method is easy to be implemented and can be used to investigate the miRNA effective regulation underlying the expression change profiles obtained from microarray experiments.
Conflict of interest statement
Figures

References
-
- Xu P, Guo M, Hay BA. MicroRNAs and the regulation of cell death. Trends Genet. 2004;20:617–624. - PubMed
-
- Karp X, Ambros V. Encountering microRNAs in cell fate signaling. Science. 2005;310:1288–1289. - PubMed
-
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. - PubMed
-
- Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

