Plasmodium falciparum antigenic variation. Mapping mosaic var gene sequences onto a network of shared, highly polymorphic sequence blocks
- PMID: 18433451
- PMCID: PMC2440560
- DOI: 10.1111/j.1365-2958.2008.06248.x
Plasmodium falciparum antigenic variation. Mapping mosaic var gene sequences onto a network of shared, highly polymorphic sequence blocks
Abstract
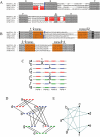
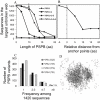
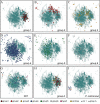
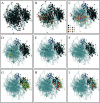
Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) is a potentially important family of immune targets, encoded by an extremely diverse gene family called var. Understanding of the genetic organization of var genes is hampered by sequence mosaicism that results from a long history of non-homologous recombination. Here we have used software designed to analyse social networks to visualize the relationships between large collections of short var sequences tags sampled from clinical parasite isolates. In this approach, two sequences are connected if they share one or more highly polymorphic sequence blocks. The results show that the majority of analysed sequences including several var-like sequences from the chimpanzee parasite Plasmodium reichenowi can be either directly or indirectly linked together in a single unbroken network. However, the network is highly structured and contains putative subgroups of recombining sequences. The major subgroup contains the previously described group A var genes, previously proposed to be genetically distinct. Another subgroup contains sequences found to be associated with rosetting, a parasite virulence phenotype. The mosaic structure of the sequences and their division into subgroups may reflect the conflicting problems of maximizing antigenic diversity and minimizing epitope sharing between variants while maintaining their host cell binding functions.
Figures


References
-
- Aguiar JC, Albrecht GR, Cegielski P, Greenwood BM, Jensen JB, Lallinger G, et al. Agglutination of Plasmodium falciparum-infected erythrocytes from east and west African isolates by human sera from distant geographical regions. Am J Trop Med Hyg. 1992;47:621–632. - PubMed
-
- Albrecht L, Merino EF, Hoffmann EH, Ferreira MU, de Mattos Ferreira RG, Osakabe AL, et al. Extense variant gene family repertoire overlap in Western Amazon Plasmodium falciparum isolates. Mol Biochem Parasitol. 2006;150:157–165. - PubMed
-
- Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, et al. Cloning the Plasmodium falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

