Rex1/Zfp42 is dispensable for pluripotency in mouse ES cells
- PMID: 18433507
- PMCID: PMC2386458
- DOI: 10.1186/1471-213X-8-45
Rex1/Zfp42 is dispensable for pluripotency in mouse ES cells
Abstract
Background: Rex1/Zfp42 has been extensively used as a marker for the undifferentiated state of pluripotent stem cells. However, its function in pluripotent stem cells including embryonic stem (ES) cells remained unclear although its involvement in visceral endoderm differentiation in F9 embryonal carcinoma (EC) cells was reported.
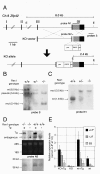
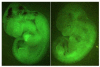
Results: We showed the function of Rex1 in mouse ES cells as well as in embryos using the conventional gene targeting strategy. Our results clearly indicated that Rex1 function is dispensable for both the maintenance of pluripotency in ES cells and the development of embryos. However, Rex1-/- ES cells showed the defect to induce a subset of the marker genes of visceral endoderm, when differentiated as embryoid body, as found in EC cells.
Conclusion: Rex1 should be regarded just as a marker of pluripotency without functional significance like the activity of alkaline phosphatase.
Figures



Similar articles
-
Demarcation of stable subpopulations within the pluripotent hESC compartment.PLoS One. 2013;8(2):e57276. doi: 10.1371/journal.pone.0057276. Epub 2013 Feb 21. PLoS One. 2013. PMID: 23437358 Free PMC article.
-
Analysis of Rex1 (zfp42) function in embryonic stem cell differentiation.Dev Dyn. 2009 Aug;238(8):1863-77. doi: 10.1002/dvdy.22037. Dev Dyn. 2009. PMID: 19618472 Free PMC article.
-
Identification and characterization of subpopulations in undifferentiated ES cell culture.Development. 2008 Mar;135(5):909-18. doi: 10.1242/dev.017400. Development. 2008. PMID: 18263842
-
Animal embryonic stem (ES) cells: self-renewal, pluripotency, transgenesis and nuclear transfer.Hum Cell. 2004 Sep;17(3):107-15. doi: 10.1111/j.1749-0774.2004.tb00026.x. Hum Cell. 2004. PMID: 15859155 Review.
-
Stem cell potency and the ability to contribute to chimeric organisms.Reproduction. 2013 Mar 7;145(3):R81-8. doi: 10.1530/REP-12-0396. Print 2013 Mar 1. Reproduction. 2013. PMID: 23221011 Free PMC article. Review.
Cited by
-
Repression and 3D-restructuring resolves regulatory conflicts in evolutionarily rearranged genomes.Cell. 2022 Sep 29;185(20):3689-3704.e21. doi: 10.1016/j.cell.2022.09.006. Cell. 2022. PMID: 36179666 Free PMC article.
-
Reduction of pluripotent gene expression in murine embryonic stem cells exposed to mechanical loading or Cyclo RGD peptide.BMC Cell Biol. 2017 Nov 14;18(1):32. doi: 10.1186/s12860-017-0148-6. BMC Cell Biol. 2017. PMID: 29137597 Free PMC article.
-
Novel kinetic and developmental transcriptomic pan-stress responses by embryonic stem cells.Sci Rep. 2025 Jul 1;15(1):21291. doi: 10.1038/s41598-025-06628-z. Sci Rep. 2025. PMID: 40594963 Free PMC article.
-
Stress Decreases Host Viral Resistance and Increases Covid Susceptibility in Embryonic Stem Cells.Stem Cell Rev Rep. 2021 Dec;17(6):2164-2177. doi: 10.1007/s12015-021-10188-w. Epub 2021 Jun 21. Stem Cell Rev Rep. 2021. PMID: 34155611 Free PMC article.
-
Demarcation of stable subpopulations within the pluripotent hESC compartment.PLoS One. 2013;8(2):e57276. doi: 10.1371/journal.pone.0057276. Epub 2013 Feb 21. PLoS One. 2013. PMID: 23437358 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

