Molecular dynamics simulation of the Escherichia coli NikR protein: equilibrium conformational fluctuations reveal interdomain allosteric communication pathways
- PMID: 18433769
- PMCID: PMC2478562
- DOI: 10.1016/j.jmb.2008.03.010
Molecular dynamics simulation of the Escherichia coli NikR protein: equilibrium conformational fluctuations reveal interdomain allosteric communication pathways
Abstract
Escherichia coli NikR is a homotetrameric Ni(2+)- and DNA-binding protein that functions as a transcriptional repressor of the NikABCDE nickel permease. The protein is composed of two distinct domains. The N-terminal 50 amino acids of each chain forms part of the dimeric ribbon-helix-helix (RHH) domains, a well-studied DNA-binding fold. The 83-residue C-terminal nickel-binding domain forms an ACT (aspartokinase, chorismate mutase, and TyrA) fold and contains the tetrameric interface. In this study, we have utilized an equilibrium molecular dynamics simulation in order to explore the conformational dynamics of the NikR tetramer and determine important residue interactions within and between the RHH and ACT domains to gain insight into the effects of Ni(2+) on DNA-binding activity. The molecular simulation data were analyzed using two different correlation measures based on fluctuations in atomic position and noncovalent contacts together with a clustering algorithm to define groups of residues with similar correlation patterns for both types of correlation measure. Based on these analyses, we have defined a series of residue interrelationships that describe an allosteric communication pathway between the Ni(2+)- and DNA-binding sites, which are separated by 40 A. Several of the residues identified by our analyses have been previously shown experimentally to be important for NikR function. An additional subset of the identified residues structurally connects the experimentally implicated residues and may help coordinate the allosteric communication between the ACT and RHH domains.
Figures
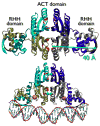







Similar articles
-
Crystal structure of the nickel-responsive transcription factor NikR.Nat Struct Biol. 2003 Oct;10(10):794-9. doi: 10.1038/nsb985. Epub 2003 Sep 14. Nat Struct Biol. 2003. PMID: 12970756
-
Nickel coordination is regulated by the DNA-bound state of NikR.Nat Struct Biol. 2003 Feb;10(2):126-30. doi: 10.1038/nsb890. Nat Struct Biol. 2003. PMID: 12524532
-
Protease digestion analysis of Escherichia coli NikR: evidence for conformational stabilization with Ni(II).J Biol Inorg Chem. 2005 Oct;10(6):605-12. doi: 10.1007/s00775-005-0008-2. Epub 2005 Nov 2. J Biol Inorg Chem. 2005. PMID: 16133200
-
Regulation of high affinity nickel uptake in bacteria. Ni2+-Dependent interaction of NikR with wild-type and mutant operator sites.J Biol Chem. 2000 Jun 30;275(26):19735-41. doi: 10.1074/jbc.M002232200. J Biol Chem. 2000. PMID: 10787413
-
Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins.Curr Issues Mol Biol. 2006 Jan;8(1):51-62. Curr Issues Mol Biol. 2006. PMID: 16450885 Review.
Cited by
-
ProSNEx: a web-based application for exploration and analysis of protein structures using network formalism.Nucleic Acids Res. 2019 Jul 2;47(W1):W471-W476. doi: 10.1093/nar/gkz390. Nucleic Acids Res. 2019. PMID: 31114881 Free PMC article.
-
Investigating dynamic and energetic determinants of protein nucleic acid recognition: analysis of the zinc finger zif268-DNA complexes.BMC Struct Biol. 2010 Nov 24;10:42. doi: 10.1186/1472-6807-10-42. BMC Struct Biol. 2010. PMID: 21106075 Free PMC article.
-
Exploring residue component contributions to dynamical network models of allostery.J Chem Theory Comput. 2012 Aug 14;8(8):2949-2961. doi: 10.1021/ct300377a. Epub 2012 Jul 5. J Chem Theory Comput. 2012. PMID: 23139645 Free PMC article.
-
Discovery of Novel Allosteric Eg5 Inhibitors Through Structure-Based Virtual Screening.Chem Biol Drug Des. 2016 Aug;88(2):178-87. doi: 10.1111/cbdd.12744. Epub 2016 Mar 6. Chem Biol Drug Des. 2016. PMID: 26864917 Free PMC article.
-
Molecular dynamics and docking simulations as a proof of high flexibility in E. coli FabH and its relevance for accurate inhibitor modeling.J Comput Aided Mol Des. 2011 Apr;25(4):371-93. doi: 10.1007/s10822-011-9427-z. Epub 2011 Apr 23. J Comput Aided Mol Des. 2011. PMID: 21516317
References
-
- Vignais PM, Billoud B, Meyer J. Classification and phylogeny of hydrogenases. FEMS Microbiol Rev. 2001;25:455–501. - PubMed
-
- Navarro C, Wu LF, Mandrand-Berthelot MA. The nik operon of Eschericia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Mol Microbiol. 1993;9:1181–1191. - PubMed
-
- Chivers PT, Sauer RT. Regulation of high affinity nickel uptake in bacteria. Ni2+-Dependent interaction of NikR with wild-type and mutant operator sites. J Biol Chem. 2000;275:19735–41. - PubMed
-
- Iwig JS, Rowe JL, Chivers PT. Nickel homeostasis in Escherichia coli – the rcnR-rcnA efflux pathway and its linkage to NikR function. Molecular Microbiology. 2006;62:252–62. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

