Side chain substitution benchmark for peptide/MHC interaction
- PMID: 18434501
- PMCID: PMC2386744
- DOI: 10.1110/ps.073402508
Side chain substitution benchmark for peptide/MHC interaction
Abstract
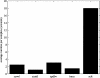
The prediction of T-cell epitopes is an essential part in virtual immunology. Apart from sequence-based techniques, which achieve good results but fail to give insight into the binding behavior of a certain peptide binding to a major histocompatibility complex, structure-based approaches are another important technique. An essential step is the correct placement of the side chains for a given peptide in cases where no experimental data for the structure are available. To our knowledge, no benchmark for side chain substitution in the area of HLA has been reported in the literature. Here, we present a comparison of five different tools (SCWRL, SCATD, SPDBV, SCit, IRECS) applicable for side chain substitution. Each tool is tested on 29 different HLA-A2 structures with experimentally known side chain positions. Parts of the benchmark are correctness, reliability, runtime, and usability. For validation, the root mean square deviations between X-ray structures and predicted structures are used. All tools show different strengths and weaknesses.
Figures

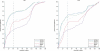


Similar articles
-
PeptX: using genetic algorithms to optimize peptides for MHC binding.BMC Bioinformatics. 2011 Jun 17;12:241. doi: 10.1186/1471-2105-12-241. BMC Bioinformatics. 2011. PMID: 21679477 Free PMC article.
-
Structural prediction of peptides binding to MHC class I molecules.Proteins. 2006 Apr 1;63(1):43-52. doi: 10.1002/prot.20870. Proteins. 2006. PMID: 16447245
-
Toward the prediction of class I and II mouse major histocompatibility complex-peptide-binding affinity: in silico bioinformatic step-by-step guide using quantitative structure-activity relationships.Methods Mol Biol. 2007;409:227-45. doi: 10.1007/978-1-60327-118-9_16. Methods Mol Biol. 2007. PMID: 18450004
-
Toward more accurate pan-specific MHC-peptide binding prediction: a review of current methods and tools.Brief Bioinform. 2012 May;13(3):350-64. doi: 10.1093/bib/bbr060. Epub 2011 Sep 22. Brief Bioinform. 2012. PMID: 21949215 Review.
-
Predicting peptide binding to Major Histocompatibility Complex molecules.Autoimmun Rev. 2011 Jun;10(8):469-73. doi: 10.1016/j.autrev.2011.02.003. Epub 2011 Feb 16. Autoimmun Rev. 2011. PMID: 21333759 Review.
Cited by
-
A critical cross-validation of high throughput structural binding prediction methods for pMHC.J Comput Aided Mol Des. 2009 May;23(5):301-7. doi: 10.1007/s10822-009-9259-2. Epub 2009 Feb 5. J Comput Aided Mol Des. 2009. PMID: 19194661
-
PeptX: using genetic algorithms to optimize peptides for MHC binding.BMC Bioinformatics. 2011 Jun 17;12:241. doi: 10.1186/1471-2105-12-241. BMC Bioinformatics. 2011. PMID: 21679477 Free PMC article.
-
Characterization of PDZ domain-peptide interactions using an integrated protocol of QM/MM, PB/SA, and CFEA analyses.J Comput Aided Mol Des. 2011 Oct;25(10):947-58. doi: 10.1007/s10822-011-9474-5. Epub 2011 Oct 1. J Comput Aided Mol Des. 2011. PMID: 21964565
-
Large scale characterization of the LC13 TCR and HLA-B8 structural landscape in reaction to 172 altered peptide ligands: a molecular dynamics simulation study.PLoS Comput Biol. 2014 Aug 7;10(8):e1003748. doi: 10.1371/journal.pcbi.1003748. eCollection 2014 Aug. PLoS Comput Biol. 2014. PMID: 25101830 Free PMC article.
-
Exploring peptide/MHC detachment processes using hierarchical natural move Monte Carlo.Bioinformatics. 2016 Jan 15;32(2):181-6. doi: 10.1093/bioinformatics/btv502. Epub 2015 Sep 22. Bioinformatics. 2016. PMID: 26395770 Free PMC article.
References
-
- Berendsen, H.J.C., van der Spoel, D., van Drunen, R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56.
-
- Bordner, A.J., Abagyan, R. Ab initio prediction of peptide-MHC binding geometry for diverse class I MHC allotypes. Proteins. 2006;63:512–526. - PubMed
-
- Bui, H.H., Schiewe, A.J., von, G.H., Haworth, I.S. Structural prediction of peptides binding to MHC class I molecules. Proteins. 2006;63:43–52. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

