Molecular architecture of Galphao and the structural basis for RGS16-mediated deactivation
- PMID: 18434540
- PMCID: PMC2359805
- DOI: 10.1073/pnas.0801569105
Molecular architecture of Galphao and the structural basis for RGS16-mediated deactivation
Abstract
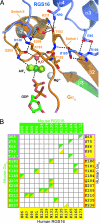
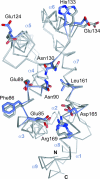
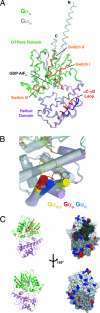
Heterotrimeric G proteins relay extracellular cues from heptahelical transmembrane receptors to downstream effector molecules. Composed of an alpha subunit with intrinsic GTPase activity and a betagamma heterodimer, the trimeric complex dissociates upon receptor-mediated nucleotide exchange on the alpha subunit, enabling each component to engage downstream effector targets for either activation or inhibition as dictated in a particular pathway. To mitigate excessive effector engagement and concomitant signal transmission, the Galpha subunit's intrinsic activation timer (the rate of GTP hydrolysis) is regulated spatially and temporally by a class of GTPase accelerating proteins (GAPs) known as the regulator of G protein signaling (RGS) family. The array of G protein-coupled receptors, Galpha subunits, RGS proteins and downstream effectors in mammalian systems is vast. Understanding the molecular determinants of specificity is critical for a comprehensive mapping of the G protein system. Here, we present the 2.9 A crystal structure of the enigmatic, neuronal G protein Galpha(o) in the GTP hydrolytic transition state, complexed with RGS16. Comparison with the 1.89 A structure of apo-RGS16, also presented here, reveals plasticity upon Galpha(o) binding, the determinants for GAP activity, and the structurally unique features of Galpha(o) that likely distinguish it physiologically from other members of the larger Galpha(i) family, affording insight to receptor, GAP and effector specificity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
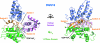
References
-
- Koelle MR, Horvitz HR. EGL-10 regulates G protein signaling in the C. elegans nervous system and shares a conserved domain with many mammalian proteins. Cell. 1996;84:115–125. - PubMed
-
- Berman DM, Wilkie TM, Gilman AG. GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein alpha subunits. Cell. 1996;86:445–452. - PubMed
-
- Berman DM, Kozasa T, Gilman AG. The GTPase-activating protein RGS4 stabilizes the transition state for nucleotide hydrolysis. J Biol Chem. 1996;271:27209–27212. - PubMed
-
- Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AlF4-activated G(i alpha1): Stabilization of the transition state for GTP hydrolysis. Cell. 1997;89:251–261. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

