Unusually rapid evolution of Neuroligin-4 in mice
- PMID: 18434543
- PMCID: PMC2359808
- DOI: 10.1073/pnas.0801383105
Unusually rapid evolution of Neuroligin-4 in mice
Abstract
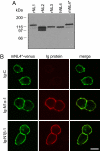
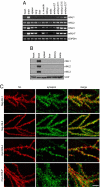
Neuroligins (NLs) are postsynaptic cell-adhesion molecules that are implicated in humans in autism spectrum disorders because the genes encoding NL3 and NL4 are mutated in rare cases of familial autism. NLs are highly conserved evolutionarily, except that no NL4 was detected in the currently available mouse genome sequence assemblies. We now demonstrate that mice express a distant NL4 variant that rapidly evolved from other mammalian NL4 genes and that exhibits sequence variations even between different mouse strains. Despite its divergence, mouse NL4 binds neurexins and is transported into dendritic spines, suggesting that the core properties of NLs are retained in this divergent NL isoform. The selectively rapid evolution of NL4 in mice suggests that its function in the brain is under less stringent control than that of other NLs, shedding light on why its mutation in autism spectrum disorder patients is not lethal, but instead leads to a discrete developmental brain disorder.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Ichtchenko K, et al. Neuroligin 1: A splice site-specific ligand for β-neurexins. Cell. 1995;81:435–443. - PubMed
-
- Ichtchenko K, Nguyen T, Südhof TC. Structures, alternative splicing, and neurexin binding of multiple neuroligins. J Biol Chem. 1996;271:2676–2682. - PubMed
-
- Ushkaryov YA, Petrenko AG, Geppert M, Südhof TC. Neurexins: Synaptic cell surface proteins related to the α-latrotoxin receptor and laminin. Science. 1992;257:50–56. - PubMed
-
- Nguyen T, Südhof TC. Binding properties of neuroligin 1 and neurexin 1β reveal function as heterophilic cell adhesion molecules. J Biol Chem. 1997;272:26032–26039. - PubMed
-
- Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

