Origins of gonadotropin-releasing hormone (GnRH) in vertebrates: identification of a novel GnRH in a basal vertebrate, the sea lamprey
- PMID: 18436713
- PMCID: PMC2488216
- DOI: 10.1210/en.2008-0184
Origins of gonadotropin-releasing hormone (GnRH) in vertebrates: identification of a novel GnRH in a basal vertebrate, the sea lamprey
Abstract
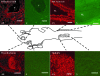
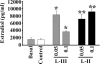
We cloned a cDNA encoding a novel (GnRH), named lamprey GnRH-II, from the sea lamprey, a basal vertebrate. The deduced amino acid sequence of the newly identified lamprey GnRH-II is QHWSHGWFPG. The architecture of the precursor is similar to that reported for other GnRH precursors consisting of a signal peptide, decapeptide, a downstream processing site, and a GnRH-associated peptide; however, the gene for lamprey GnRH-II does not have introns in comparison with the gene organization for all other vertebrate GnRHs. Lamprey GnRH-II precursor transcript was widely expressed in a variety of tissues. In situ hybridization of the brain showed expression and localization of the transcript in the hypothalamus, medulla, and olfactory regions, whereas immunohistochemistry using a specific antiserum showed only GnRH-II cell bodies and processes in the preoptic nucleus/hypothalamus areas. Lamprey GnRH-II was shown to stimulate the hypothalamic-pituitary axis using in vivo and in vitro studies. Lamprey GnRH-II was also shown to activate the inositol phosphate signaling system in COS-7 cells transiently transfected with the lamprey GnRH receptor. These studies provide evidence for a novel lamprey GnRH that has a role as a third hypothalamic GnRH. In summary, the newly discovered lamprey GnRH-II offers a new paradigm of the origin of the vertebrate GnRH family. We hypothesize that due to a genome/gene duplication event, an ancestral gene gave rise to two lineages of GnRHs: the gnathostome GnRH and lamprey GnRH-II.
Figures






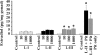
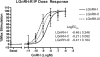

Similar articles
-
Two novel gonadotropin-releasing hormones (GnRHs) from the urochordate ascidian, Halocynthia roretzi: implications for the origin of vertebrate GnRH isoforms.Zoolog Sci. 2013 Apr;30(4):311-8. doi: 10.2108/zsj.30.311. Zoolog Sci. 2013. PMID: 23537242
-
Identification of sea lamprey GTHbeta-like cDNA and its evolutionary implications.Gen Comp Endocrinol. 2006 Aug;148(1):22-32. doi: 10.1016/j.ygcen.2005.11.009. Epub 2006 Jan 19. Gen Comp Endocrinol. 2006. PMID: 16427051
-
Cloning and analysis of the lamprey GnRH-III cDNA from eight species of lamprey representing the three families of Petromyzoniformes.Gen Comp Endocrinol. 2004 Oct;139(1):85-94. doi: 10.1016/j.ygcen.2004.07.011. Gen Comp Endocrinol. 2004. PMID: 15474539
-
The origins of the vertebrate hypothalamic-pituitary-gonadal (HPG) and hypothalamic-pituitary-thyroid (HPT) endocrine systems: new insights from lampreys.Gen Comp Endocrinol. 2009 Mar;161(1):20-9. doi: 10.1016/j.ygcen.2008.11.023. Epub 2008 Dec 3. Gen Comp Endocrinol. 2009. PMID: 19084529 Review.
-
Landmark discoveries in elucidating the origins of the hypothalamic-pituitary system from the perspective of a basal vertebrate, sea lamprey.Gen Comp Endocrinol. 2018 Aug 1;264:3-15. doi: 10.1016/j.ygcen.2017.10.016. Epub 2017 Oct 27. Gen Comp Endocrinol. 2018. PMID: 29111305 Review.
Cited by
-
Molecular Coevolution of Neuropeptides Gonadotropin-Releasing Hormone and Kisspeptin with their Cognate G Protein-Coupled Receptors.Front Neurosci. 2012 Jan 24;6:3. doi: 10.3389/fnins.2012.00003. eCollection 2012. Front Neurosci. 2012. PMID: 22291614 Free PMC article.
-
The interrelationship of estrogen receptor and GnRH in a Basal vertebrate, the sea lamprey.Front Endocrinol (Lausanne). 2011 Oct 28;2:58. doi: 10.3389/fendo.2011.00058. eCollection 2011. Front Endocrinol (Lausanne). 2011. PMID: 22654815 Free PMC article.
-
Iso-petromyroxols: novel dihydroxylated tetrahydrofuran enantiomers from sea lamprey (Petromyzon marinus).Molecules. 2015 Mar 23;20(3):5215-22. doi: 10.3390/molecules20035215. Molecules. 2015. PMID: 25806547 Free PMC article.
-
Expression of polyamines and its association with GnRH-I in the hypothalamus during aging in rodent model.Amino Acids. 2022 Aug;54(8):1135-1154. doi: 10.1007/s00726-022-03139-3. Epub 2022 Mar 14. Amino Acids. 2022. PMID: 35286462
-
Molecular cloning and expression of caspase-3 in the protandrous cinnamon clownfish, Amphiprion melanopus, during sex change.Fish Physiol Biochem. 2013 Jun;39(3):417-29. doi: 10.1007/s10695-012-9709-y. Epub 2012 Aug 29. Fish Physiol Biochem. 2013. PMID: 22926760
References
-
- Matsuo H, Baba Y, Nair RM, Arimura A, Schally AV 1971 Structure of the porcine LH- and FSH-releasing hormone. I. The proposed amino acid sequence. Biochem Biophys Res Commun 43:1334–1339 - PubMed
-
- Kah O, Lethimonier C, Somoza G, Guilgur LG, Vaillant C, Lareyre JJ 2007 GnRH and GnRH receptors in metazoa: a historical, comparative, and evolutive perspective. Gen Comp Endocrinol 153:346–364 - PubMed
-
- Silver MR, Kawauchi H, Nozaki M, Sower SA 2004 Cloning and analysis of the lamprey GnRH-III cDNA from eight species of lamprey representing the three families of Petromyzoniformes. Gen Comp Endocrinol 139:85–94 - PubMed

