Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China
- PMID: 18439357
- PMCID: PMC2600263
- DOI: 10.3201/eid1405.071312
Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China
Abstract
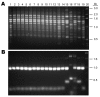
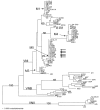
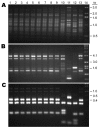
To determine the population structure of the cryptococcosis agents in China, we analyzed the genotype of 120 Cryptococcus neoformans and 9 Cryptococcus gattii strains isolated from 1980 through 2006 from cryptococcosis patients residing in 16 provinces of mainland China. A total of 71% (91/129) of the clinical strains isolated from 1985 through 2006 were from patients without any apparent risk factors. Only 8.5% (11/129) were from AIDS patients; the remaining 20.5% (27/129) were from patients with underlying diseases other than HIV infection. One hundred twenty of the 129 isolates were C. neoformans serotype A, mating type MATalpha strains that exhibited an identical M13-based VNI subtype, which was distinguishable from the reference VNI molecular type. The 9 remaining isolates were serotype B, MATalpha strains of C. gattii and portrayed a typical VGI molecular type. Data analyzed from multilocus sequences showed no variation and that these Chinese C. neoformans isolates belong to a cluster that has phylogenetically diverged from the VNI reference strain. Our finding that most cryptococcosis patients in China had no apparent risk factor is in stark contrast with reports from other countries.
Figures



References
-
- Kwon-Chung KJ, Bennett JE. Medical mycology. Philadelphia: Lea & Febiger; 1992.
-
- Litvintseva AP, Thakur R, Reller LB, Mitchell TG. Prevalence of clinical isolates of Cryptococcus gattii serotype C among patients with AIDS in sub-Saharan Africa. Eukaryot Cell. 2005;192:888–92. - PubMed
-
- Meyer W, Marszewska K, Amirmostofian M, Igreja RP, Hardtke C, Methling K, et al. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA—a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis. 1999;20:1790–9. 10.1002/(SICI)1522-2683(19990101)20:8<1790::AID-ELPS1790>3.0.CO;2-2 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
