Tumor necrosis factor-alpha and endothelial cells modulate Notch signaling in the bone marrow microenvironment during inflammation
- PMID: 18439488
- PMCID: PMC3437760
- DOI: 10.1016/j.exphem.2007.12.012
Tumor necrosis factor-alpha and endothelial cells modulate Notch signaling in the bone marrow microenvironment during inflammation
Abstract
Objective: Homeostasis of the hematopoietic compartment is challenged and maintained during conditions of stress by mechanisms that are poorly defined. To understand how the bone marrow (BM) microenvironment influences hematopoiesis, we explored the role of Notch signaling and BM endothelial cells in providing microenvironmental cues to hematopoietic cells in the presence of inflammatory stimuli.
Materials and methods: The human BM endothelial cell line (BMEC) and primary human BM endothelial cells were analyzed for expression of Notch ligands and the ability to expand hematopoietic progenitors in an in vitro coculture system. In vivo experiments were carried out to identify modulation of Notch signaling in BM endothelial and hematopoietic cells in mice challenged with tumor necrosis factor-alpha (TNF-alpha) or lipopolysaccharide (LPS), or in Tie2-tmTNF-alpha transgenic mice characterized by constitutive TNF-alpha activation.
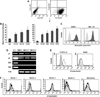
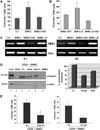
Results: BM endothelial cells were found to express Jagged ligands and to greatly support progenitor's colony-forming ability. This effect was markedly decreased by Notch antagonists and augmented by increasing levels of Jagged2. Physiologic upregulation of Jagged2 expression on BMEC was observed upon TNF-alpha activation. Injection of TNF-alpha or LPS upregulated three- to fourfold Jagged2 expression on murine BM endothelial cells in vivo and resulted in increased Notch activation on murine hematopoietic stem/progenitor cells. Similarly, constitutive activation of endothelial cells in Tie2-tmTNF-alpha mice was characterized by increased expression of Jagged2 and by augmented Notch activation on hematopoietic stem/progenitor cells.
Conclusions: Our results provide the first evidence that BM endothelial cells promote expansion of hematopoietic progenitor cells by a Notch-dependent mechanism and that TNF-alpha and LPS can modulate the levels of Notch ligand expression and Notch activation in the BM microenvironment in vivo.
Figures




Similar articles
-
The Notch ligands Jagged2, Delta1, and Delta4 induce differentiation and expansion of functional human NK cells from CD34+ cord blood hematopoietic progenitor cells.Biol Blood Marrow Transplant. 2009 Sep;15(9):1026-37. doi: 10.1016/j.bbmt.2009.06.002. Biol Blood Marrow Transplant. 2009. PMID: 19660715 Free PMC article.
-
Jagged2 acts as a Delta-like Notch ligand during early hematopoietic cell fate decisions.Blood. 2011 Apr 28;117(17):4449-59. doi: 10.1182/blood-2010-06-290049. Epub 2011 Mar 3. Blood. 2011. PMID: 21372153 Free PMC article.
-
Hypoxia-induced Jagged2 promotes breast cancer metastasis and self-renewal of cancer stem-like cells.Oncogene. 2011 Sep 29;30(39):4075-86. doi: 10.1038/onc.2011.122. Epub 2011 Apr 18. Oncogene. 2011. PMID: 21499308 Free PMC article.
-
Jagged2-expressing hematopoietic progenitors promote regulatory T cell expansion in the periphery through notch signaling.Immunity. 2006 Nov;25(5):823-34. doi: 10.1016/j.immuni.2006.09.008. Epub 2006 Nov 2. Immunity. 2006. PMID: 17081781
-
Recent advances in hematopoietic stem cell biology.Curr Opin Hematol. 2004 Nov;11(6):392-8. doi: 10.1097/01.moh.0000145672.42503.70. Curr Opin Hematol. 2004. PMID: 15548993 Review.
Cited by
-
Diversity of Vascular Niches in Bones and Joints During Homeostasis, Ageing, and Diseases.Front Immunol. 2021 Dec 17;12:798211. doi: 10.3389/fimmu.2021.798211. eCollection 2021. Front Immunol. 2021. PMID: 34975909 Free PMC article. Review.
-
Bone marrow mesenchymal stem cells promote remyelination in spinal cord by driving oligodendrocyte progenitor cell differentiation via TNFα/RelB-Hes1 pathway: a rat model study of 2,5-hexanedione-induced neurotoxicity.Stem Cell Res Ther. 2021 Aug 4;12(1):436. doi: 10.1186/s13287-021-02518-z. Stem Cell Res Ther. 2021. PMID: 34348774 Free PMC article.
-
Proinflammatory signaling regulates hematopoietic stem cell emergence.Cell. 2014 Nov 20;159(5):1070-1085. doi: 10.1016/j.cell.2014.10.031. Epub 2014 Nov 6. Cell. 2014. PMID: 25416946 Free PMC article.
-
Notch Signaling in the Regulation of Hematopoietic Stem Cell.Curr Stem Cell Rep. 2017;3(3):202-209. doi: 10.1007/s40778-017-0090-8. Epub 2017 Jul 10. Curr Stem Cell Rep. 2017. PMID: 28845387 Free PMC article. Review.
-
PathExpSurv: pathway expansion for explainable survival analysis and disease gene discovery.BMC Bioinformatics. 2023 Nov 15;24(1):434. doi: 10.1186/s12859-023-05535-2. BMC Bioinformatics. 2023. PMID: 37968615 Free PMC article.
References
-
- Suda T, Arai F, Hirao A. Hematopoietic stem cells and their niche. Trends Immunol. 2005;26:426–433. - PubMed
-
- Rafii S, Shapiro F, Pettengell R, et al. Human bone marrow microvascular endothelial cells support long-term proliferation and differentiation of myeloid and megakaryocytic progenitors. Blood. 1995;86:3353–3363. - PubMed
-
- Cines DB, Pollak ES, Buck CA, et al. Endothelial cells in physiology and in the pathophysiology of vascular disorders. Blood. 1998;91:3527–3561. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

