Trypanosoma brucei RNA editing protein TbMP42 (band VI) is crucial for the endonucleolytic cleavages but not the subsequent steps of U-deletion and U-insertion
- PMID: 18441050
- PMCID: PMC2390806
- DOI: 10.1261/rna.899508
Trypanosoma brucei RNA editing protein TbMP42 (band VI) is crucial for the endonucleolytic cleavages but not the subsequent steps of U-deletion and U-insertion
Abstract
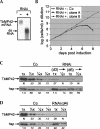
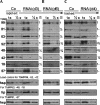
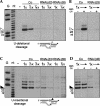
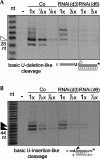
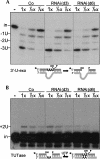
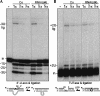
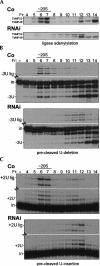
Trypanosome mitochondrial mRNAs achieve their coding sequences through RNA editing. This process, catalyzed by approximately 20S protein complexes, involves large numbers of uridylate (U) insertions and deletions within mRNA precursors. Here we analyze the role of the essential TbMP42 protein (band VI/KREPA2) by individually examining each step of the U-deletional and U-insertional editing cycles, using reactions in the approximately linear range. We examined control extracts and RNA interference (RNAi) extracts prepared soon after TbMP42 was depleted (when primary effects should be most evident) and three days later (when precedent shows secondary effects can become prominent). This analysis shows TbMP42 is critical for cleavage of editing substrates by both the U-deletional and U-insertional endonucleases. However, on simple substrates that assess cleavage independent of editing features, TbMP42 is similarly required only for the U-deletional endonuclease, indicating TbMP42 affects the two editing endonucleases differently. Supplementing RNAi extract with recombinant TbMP42 partly restores these cleavage activities. Notably, we find that all the other editing steps (the 3'-U-exonuclease [3'-U-exo] and ligation steps of U-deletion and the terminal-U-transferase [TUTase] and ligation steps of U-insertion) remain at control levels upon RNAi induction, and hence are not dependent on TbMP42. This contrasts with an earlier report that TbMP42 is a 3'-U-exo that may act in U-deletion and additionally is critical for the TUTase and/or ligation steps of U-insertion, observations our data suggest reflect indirect effects of TbMP42 depletion. Thus, trypanosomes require TbMP42 for both endonucleolytic cleavage steps of RNA editing, but not for any of the subsequent steps of the editing cycles.
Figures



Similar articles
-
In Trypanosoma brucei RNA editing, TbMP18 (band VII) is critical for editosome integrity and for both insertional and deletional cleavages.Mol Cell Biol. 2007 Jan;27(2):777-87. doi: 10.1128/MCB.01460-06. Epub 2006 Nov 13. Mol Cell Biol. 2007. PMID: 17101787 Free PMC article.
-
In Trypanosoma brucei RNA editing, band II enables recognition specifically at each step of the U insertion cycle.Mol Cell Biol. 2005 Apr;25(7):2785-94. doi: 10.1128/MCB.25.7.2785-2794.2005. Mol Cell Biol. 2005. PMID: 15767682 Free PMC article.
-
T. brucei RNA editing: action of the U-insertional TUTase within a U-deletion cycle.RNA. 2006 Mar;12(3):476-87. doi: 10.1261/rna.2243206. RNA. 2006. PMID: 16495238 Free PMC article.
-
Insertional and deletional RNA editing in trypanosome mitochondria.Nucleic Acids Symp Ser. 1997;(36):15-8. Nucleic Acids Symp Ser. 1997. PMID: 9478193 Review.
-
Mitochondrial RNA editing in trypanosomes: small RNAs in control.Biochimie. 2014 May;100:125-31. doi: 10.1016/j.biochi.2014.01.003. Epub 2014 Jan 17. Biochimie. 2014. PMID: 24440637 Free PMC article. Review.
Cited by
-
The Architecture of Trypanosoma brucei editosomes.Proc Natl Acad Sci U S A. 2016 Oct 18;113(42):E6476-E6485. doi: 10.1073/pnas.1610177113. Epub 2016 Oct 5. Proc Natl Acad Sci U S A. 2016. PMID: 27708162 Free PMC article.
-
Multiple domains of the integral KREPA3 protein are critical for the structure and precise functions of RNA editing catalytic complexes in Trypanosoma brucei.RNA. 2023 Oct;29(10):1591-1609. doi: 10.1261/rna.079691.123. Epub 2023 Jul 20. RNA. 2023. PMID: 37474258 Free PMC article.
-
Inhibitors of RNA editing as potential chemotherapeutics against trypanosomatid pathogens.Int J Parasitol Drugs Drug Resist. 2011 Nov 13;2:36-46. doi: 10.1016/j.ijpddr.2011.10.003. eCollection 2012 Dec. Int J Parasitol Drugs Drug Resist. 2011. PMID: 24533263 Free PMC article. Review.
-
Kinetoplastid RNA editing involves a 3' nucleotidyl phosphatase activity.Nucleic Acids Res. 2009 Apr;37(6):1897-906. doi: 10.1093/nar/gkp049. Epub 2009 Feb 3. Nucleic Acids Res. 2009. PMID: 19190092 Free PMC article.
-
Differential Editosome Protein Function between Life Cycle Stages of Trypanosoma brucei.J Biol Chem. 2015 Oct 9;290(41):24914-31. doi: 10.1074/jbc.M115.669432. Epub 2015 Aug 24. J Biol Chem. 2015. PMID: 26304125 Free PMC article.
References
-
- Aphasizhev R., Sbicego S., Peris M., Jang S.H., Aphasizheva I., Simpson A.M., Rivlin A., Simpson L. Trypanosome mitochondrial 3′ terminal uridylyl transferase (TUTase): The key enzyme in U-insertion/deletion RNA editing. Cell. 2002;108:637–648. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
