The genome of Heliobacterium modesticaldum, a phototrophic representative of the Firmicutes containing the simplest photosynthetic apparatus
- PMID: 18441057
- PMCID: PMC2446807
- DOI: 10.1128/JB.00299-08
The genome of Heliobacterium modesticaldum, a phototrophic representative of the Firmicutes containing the simplest photosynthetic apparatus
Abstract
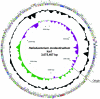
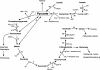
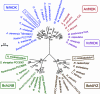
Despite the fact that heliobacteria are the only phototrophic representatives of the bacterial phylum Firmicutes, genomic analyses of these organisms have yet to be reported. Here we describe the complete sequence and analysis of the genome of Heliobacterium modesticaldum, a thermophilic species belonging to this unique group of phototrophs. The genome is a single 3.1-Mb circular chromosome containing 3,138 open reading frames. As suspected from physiological studies of heliobacteria that have failed to show photoautotrophic growth, genes encoding enzymes for known autotrophic pathways in other phototrophic organisms, including ribulose bisphosphate carboxylase (Calvin cycle), citrate lyase (reverse citric acid cycle), and malyl coenzyme A lyase (3-hydroxypropionate pathway), are not present in the H. modesticaldum genome. Thus, heliobacteria appear to be the only known anaerobic anoxygenic phototrophs that are not capable of autotrophy. Although for some cellular activities, such as nitrogen fixation, there is a full complement of genes in H. modesticaldum, other processes, including carbon metabolism and endosporulation, are more genetically streamlined than they are in most other low-G+C gram-positive bacteria. Moreover, several genes encoding photosynthetic functions in phototrophic purple bacteria are not present in the heliobacteria. In contrast to the nutritional flexibility of many anoxygenic phototrophs, the complete genome sequence of H. modesticaldum reveals an organism with a notable degree of metabolic specialization and genomic reduction.
Figures

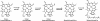
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. - PubMed
-
- Amesz, J. 1995. The antenna-reaction center complex of heliobacteria, p. 687-697. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
-
- Asao, M., D. O. Jung, L. A. Achenbach, and M. T. Madigan. 2006. Heliorestis convoluta sp. nov., a coiled, alkaliphilic heliobacterium from the Wadi El Natroun, Egypt. Extremophiles 10403-410. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

