Assembly of viral metagenomes from yellowstone hot springs
- PMID: 18441115
- PMCID: PMC2446518
- DOI: 10.1128/AEM.02598-07
Assembly of viral metagenomes from yellowstone hot springs
Abstract
Thermophilic viruses were reported decades ago; however, knowledge of their diversity, biology, and ecological impact is limited. Previous research on thermophilic viruses focused on cultivated strains. This study examined metagenomic profiles of viruses directly isolated from two mildly alkaline hot springs, Bear Paw (74 degrees C) and Octopus (93 degrees C). Using a new method for constructing libraries from picograms of DNA, nearly 30 Mb of viral DNA sequence was determined. In contrast to previous studies, sequences were assembled at 50% and 95% identity, creating composite contigs up to 35 kb and facilitating analysis of the inherent heterogeneity in the populations. Lowering the assembly identity reduced the estimated number of viral types from 1,440 and 1,310 to 548 and 283, respectively. Surprisingly, the diversity of viral species in these springs approaches that in moderate-temperature environments. While most known thermophilic viruses have a chronic, nonlytic infection lifestyle, analysis of coding sequences suggests lytic viruses are more common in geothermal environments than previously thought. The 50% assembly included one contig with high similarity and perfect synteny to nine genes from Pyrobaculum spherical virus (PSV). In fact, nearly all the genes of the 28-kb genome of PSV have apparent homologs in the metagenomes. Similarities to thermoacidophilic viruses isolated on other continents were limited to specific open reading frames but were equally strong. Nearly 25% of the reads showed significant similarity between the hot springs, suggesting a common subterranean source. To our knowledge, this is the first application of metagenomics to viruses of geothermal origin.
Figures
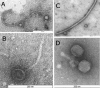
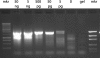
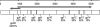
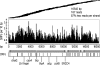

References
-
- Ahn, D. G., S. I. Kim, J. K. Rhee, K. P. Kim, J. G. Pan, and J. W. Oh. 2006. TTSV1, a new virus-like particle isolated from the hyperthermophilic crenarchaeote Thermoproteus tenax. Virology 351:280-290. - PubMed
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Angly, F. E., B. Felts, M. Breitbart, P. Salamon, R. A. Edwards, C. Carlson, A. M. Chan, M. Haynes, S. Kelley, H. Liu, J. M. Mahaffy, J. E. Mueller, J. Nulton, R. Olson, R. Parsons, S. Rayhawk, C. A. Suttle, and F. Rohwer. 2006. The marine viromes of four oceanic regions. PLoS Biol. 4:e368. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

