Functional and physiological characterization of Arabidopsis INOSITOL TRANSPORTER1, a novel tonoplast-localized transporter for myo-inositol
- PMID: 18441213
- PMCID: PMC2390729
- DOI: 10.1105/tpc.107.055632
Functional and physiological characterization of Arabidopsis INOSITOL TRANSPORTER1, a novel tonoplast-localized transporter for myo-inositol
Abstract
Arabidopsis thaliana INOSITOL TRANSPORTER1 (INT1) is a member of a small gene family with only three more genes (INT2 to INT4). INT2 and INT4 were shown to encode plasma membrane-localized transporters for different inositol epimers, and INT3 was characterized as a pseudogene. Here, we present the functional and physiological characterization of the INT1 protein, analyses of the tissue-specific expression of the INT1 gene, and analyses of phenotypic differences observed between wild-type plants and mutant lines carrying the int1.1 and int1.2 alleles. INT1 is a ubiquitously expressed gene, and Arabidopsis lines with T-DNA insertions in INT1 showed increased intracellular myo-inositol concentrations and reduced root growth. In Arabidopsis, tobacco (Nicotiana tabacum), and Saccharomyces cerevisiae, fusions of the green fluorescent protein to the C terminus of INT1 were targeted to the tonoplast membranes. Finally, patch-clamp analyses were performed on vacuoles from wild-type plants and from both int1 mutant lines to study the transport properties of INT1 at the tonoplast. In summary, the presented molecular, physiological, and functional studies demonstrate that INT1 is a tonoplast-localized H(+)/inositol symporter that mediates the efflux of inositol that is generated during the degradation of inositol-containing compounds in the vacuolar lumen.
Figures

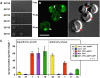


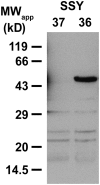



References
-
- Abel, S., and Theologis, A. (1994). Transient transformation of Arabidopsis leaf protoplasts: A versatile experimental system to study gene expression. Plant J. 5 421–427. - PubMed
-
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. - PubMed
-
- Becker, D., Kemper, E., Schell, J., and Masterson, R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20 8123–8128. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

