OsBIRH1, a DEAD-box RNA helicase with functions in modulating defence responses against pathogen infection and oxidative stress
- PMID: 18441339
- PMCID: PMC2413282
- DOI: 10.1093/jxb/ern072
OsBIRH1, a DEAD-box RNA helicase with functions in modulating defence responses against pathogen infection and oxidative stress
Abstract
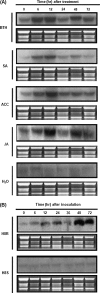
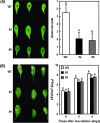
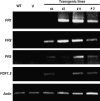
DEAD-box proteins comprise a large protein family with members from all kingdoms and play important roles in all types of processes in RNA metabolism. In this study, a rice gene OsBIRH1, which encodes a DEAD-box RNA helicase protein, was cloned and characterized. The predicted OsBIRH1 protein contains a DEAD domain and all conserved motifs that are common characteristics of DEAD-box RNA helicases. Recombinant OsBIRH1 protein purified from Escherichia coli was shown to have both RNA-dependent ATPase and ATP-dependent RNA helicase activities in vitro. Expression of OsBIRH1 was activated in rice seedling leaves after treatment with defence-related signal chemicals, for example benzothiadiazole, salicylic acid, l-aminocyclopropane-1-carboxylic acid, and jasmonic acid, and was also up-regulated in an incompatible interaction between a resistant rice genotype and the blast fungus, Magnaporthe grisea. Transgenic Arabidopsis plants that overexpress the OsBIRH1 gene were generated. Disease resistance phenotype assays revealed that the OsBIRH1-overexpressing transgenic plants showed an enhanced disease resistance against Alternaria brassicicola and Pseudomonas syringae pv. tomato DC3000. Meanwhile, defence-related genes, for example PR-1, PR-2, PR-5, and PDF1.2, showed an up-regulated expression in the transgenic plants. Moreover, the OsBIRH1 transgenic Arabidopsis plants also showed increased tolerance to oxidative stress and elevated expression levels of oxidative defence genes, AtApx1, AtApx2, and AtFSD1. The results suggest that OsBIRH1 encodes a functional DEAD-box RNA helicase and plays important roles in defence responses against biotic and abiotic stresses.
Figures





Similar articles
-
Functional analysis reveals pleiotropic effects of rice RING-H2 finger protein gene OsBIRF1 on regulation of growth and defense responses against abiotic and biotic stresses.Plant Mol Biol. 2008 Sep;68(1-2):17-30. doi: 10.1007/s11103-008-9349-x. Epub 2008 May 22. Plant Mol Biol. 2008. PMID: 18496756
-
A rice serine carboxypeptidase-like gene OsBISCPL1 is involved in regulation of defense responses against biotic and oxidative stress.Gene. 2008 Aug 15;420(1):57-65. doi: 10.1016/j.gene.2008.05.006. Epub 2008 May 20. Gene. 2008. PMID: 18571878
-
Screening for resistance against Pseudomonas syringae in rice-FOX Arabidopsis lines identified a putative receptor-like cytoplasmic kinase gene that confers resistance to major bacterial and fungal pathogens in Arabidopsis and rice.Plant Biotechnol J. 2011 May;9(4):466-85. doi: 10.1111/j.1467-7652.2010.00568.x. Epub 2010 Oct 18. Plant Biotechnol J. 2011. PMID: 20955180 Free PMC article.
-
DEAD box helicases as promising molecular tools for engineering abiotic stress tolerance in plants.Crit Rev Biotechnol. 2019 May;39(3):395-407. doi: 10.1080/07388551.2019.1566204. Epub 2019 Feb 3. Crit Rev Biotechnol. 2019. PMID: 30714414 Review.
-
Against the grain: safeguarding rice from rice blast disease.Trends Biotechnol. 2009 Mar;27(3):141-50. doi: 10.1016/j.tibtech.2008.12.002. Epub 2009 Jan 31. Trends Biotechnol. 2009. PMID: 19187990 Review.
Cited by
-
H2O2 Induces Association of RCA with the Thylakoid Membrane to Enhance Resistance of Oryza meyeriana to Xanthomonas oryzae pv. oryzae.Plants (Basel). 2019 Sep 16;8(9):351. doi: 10.3390/plants8090351. Plants (Basel). 2019. PMID: 31527548 Free PMC article.
-
Phenotypic and microarray analysis reveals salinity stress-induced oxidative tolerance in transgenic rice expressing a DEAD-box RNA helicase, OsDB10.Plant Mol Biol. 2023 Oct;113(1-3):19-32. doi: 10.1007/s11103-023-01372-2. Epub 2023 Jul 31. Plant Mol Biol. 2023. PMID: 37523054
-
Comparative Analysis Based on Transcriptomics and Metabolomics Data Reveal Differences between Emmer and Durum Wheat in Response to Nitrogen Starvation.Int J Mol Sci. 2021 Apr 30;22(9):4790. doi: 10.3390/ijms22094790. Int J Mol Sci. 2021. PMID: 33946478 Free PMC article.
-
Identification of BZR1-interacting proteins as potential components of the brassinosteroid signaling pathway in Arabidopsis through tandem affinity purification.Mol Cell Proteomics. 2013 Dec;12(12):3653-65. doi: 10.1074/mcp.M113.029256. Epub 2013 Sep 9. Mol Cell Proteomics. 2013. PMID: 24019147 Free PMC article.
-
Long-term boron-deficiency-responsive genes revealed by cDNA-AFLP differ between Citrus sinensis roots and leaves.Front Plant Sci. 2015 Jul 28;6:585. doi: 10.3389/fpls.2015.00585. eCollection 2015. Front Plant Sci. 2015. PMID: 26284101 Free PMC article.
References
-
- Arciga-Reyes L, Wootton L, Kieffer M, Davies B. UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. The Plant Journal. 2006;47:480–489. - PubMed
-
- Bartels D, Sunkar R. Drought and salt tolerance in plants. CRC Critical Reviews in Plant Sciences. 2005;24:23–58.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

