Fusion core structure of the severe acute respiratory syndrome coronavirus (SARS-CoV): in search of potent SARS-CoV entry inhibitors
- PMID: 18442051
- PMCID: PMC7166385
- DOI: 10.1002/jcb.21790
Fusion core structure of the severe acute respiratory syndrome coronavirus (SARS-CoV): in search of potent SARS-CoV entry inhibitors
Abstract
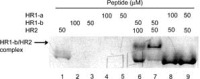
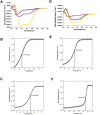
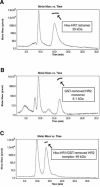
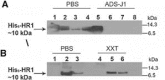
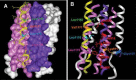
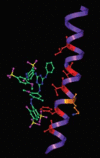
Severe acute respiratory coronavirus (SARS-CoV) spike (S) glycoprotein fusion core consists of a six-helix bundle with the three C-terminal heptad repeat (HR2) helices packed against a central coiled-coil of the other three N-terminal heptad repeat (HR1) helices. Each of the three peripheral HR2 helices shows prominent contacts with the hydrophobic surface of the central HR1 coiled-coil. The concerted protein-protein interactions among the HR helices are responsible for the fusion event that leads to the release of the SARS-CoV nucleocapsid into the target host-cell. In this investigation, we applied recombinant protein and synthetic peptide-based biophysical assays to characterize the biological activities of the HR helices. In a parallel experiment, we employed a HIV-luc/SARS pseudotyped virus entry inhibition assay to screen for potent inhibitory activities on HR peptides derived from the SARS-CoV S protein HR regions and a series of other small-molecule drugs. Three HR peptides and five small-molecule drugs were identified as potential inhibitors. ADS-J1, which has been used to interfere with the fusogenesis of HIV-1 onto CD4+ cells, demonstrated the highest HIV-luc/SARS pseudotyped virus-entry inhibition activity among the other small-molecule drugs. Molecular modeling analysis suggested that ADS-J1 may bind to the deep pocket of the hydrophobic groove on the surface of the central coiled-coil of SARS-CoV S HR protein and prevent the entrance of the SARS-CoV into the host cells.
Figures

Similar articles
-
Characterization of the heptad repeat regions, HR1 and HR2, and design of a fusion core structure model of the spike protein from severe acute respiratory syndrome (SARS) coronavirus.Biochemistry. 2004 Nov 9;43(44):14064-71. doi: 10.1021/bi049101q. Biochemistry. 2004. PMID: 15518555
-
Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors.Lancet. 2004 Mar 20;363(9413):938-47. doi: 10.1016/S0140-6736(04)15788-7. Lancet. 2004. PMID: 15043961 Free PMC article.
-
Suppression of SARS-CoV entry by peptides corresponding to heptad regions on spike glycoprotein.Biochem Biophys Res Commun. 2004 Jul 2;319(3):746-52. doi: 10.1016/j.bbrc.2004.05.046. Biochem Biophys Res Commun. 2004. PMID: 15184046 Free PMC article.
-
[Development of peptidic MERS-CoV entry inhibitors].Yao Xue Xue Bao. 2015 Dec;50(12):1513-9. Yao Xue Xue Bao. 2015. PMID: 27169270 Review. Chinese.
-
Structure and Function of the SARS-CoV-2 6-HB Fusion Core and Peptide-based Fusion Inhibitors: A Review.Curr Med Chem. 2025;32(13):2524-2546. doi: 10.2174/0109298673265694231113061842. Curr Med Chem. 2025. PMID: 38018192 Review.
Cited by
-
Inhibitors of SARS-CoV-2 Entry: Current and Future Opportunities.J Med Chem. 2020 Nov 12;63(21):12256-12274. doi: 10.1021/acs.jmedchem.0c00502. Epub 2020 Jun 25. J Med Chem. 2020. PMID: 32539378 Free PMC article. Review.
-
Cryo-EM analysis of the post-fusion structure of the SARS-CoV spike glycoprotein.Nat Commun. 2020 Jul 17;11(1):3618. doi: 10.1038/s41467-020-17371-6. Nat Commun. 2020. PMID: 32681106 Free PMC article.
-
Peptide-Based Inhibitors for SARS-CoV-2 and SARS-CoV.Adv Ther (Weinh). 2021 Oct;4(10):2100104. doi: 10.1002/adtp.202100104. Epub 2021 Aug 6. Adv Ther (Weinh). 2021. PMID: 34514085 Free PMC article. Review.
-
Antiviral activity of selected antimicrobial peptides against vaccinia virus.Antiviral Res. 2010 Jun;86(3):306-11. doi: 10.1016/j.antiviral.2010.03.012. Epub 2010 Mar 27. Antiviral Res. 2010. PMID: 20347875 Free PMC article.
-
3-Hydroxyphthalic Anhydride-Modified Chicken Ovalbumin as a Potential Candidate Inhibits SARS-CoV-2 Infection by Disrupting the Interaction of Spike Protein With Host ACE2 Receptor.Front Pharmacol. 2021 Jan 14;11:603830. doi: 10.3389/fphar.2020.603830. eCollection 2020. Front Pharmacol. 2021. PMID: 33519467 Free PMC article.
References
-
- Chim SS, Tsui SK, Chan KC, Au TC, Hung EC, Tong YK, Chiu RW, Ng EK, Chan PK, Chu CM, Sung JJ, Tam JS, Fung KP, Waye MM, Lee CY, Yuen KY, Lo YM. 2003. Genomic characterisation of the severe acute respiratory syndrome coronavirus of Amoy Gardens outbreak in Hong Kong. Lancet 362: 1807–1808. - PMC - PubMed
-
- Choy WY, Lin SG, Chan PK, Tam JS, Lo YM, Chu IM, Tsai SN, Zhong MQ, Fung KP, Waye MM, Tsui SK, Ng KO, Shan ZX, Yang M, Wu YL, Lin ZY, Ngai SM. 2004. Synthetic peptide studies on the severe acute respiratory syndrome (SARS) coronavirus spike glycoprotein: Perspective for SARS vaccine development. Clin Chem 50: 1036–1042. - PMC - PubMed
-
- Chu LH, Choy WY, Tsai SN, Rao Z, Ngai SM. 2006. Rapid peptide‐based screening on the substrate specificity of severe acute respiratory syndrome (SARS) coronavirus 3C‐like protease by matrix‐assisted laser desorption/ionization time‐of‐flight mass spectrometry. Protein Sci 15: 699–709. - PMC - PubMed
-
- Connor RI, Chen BK, Choe S, Landau NR. 1995. Vpr is required for efficient replication of human immunodeficiency virus type‐1 in mononuclear phagocytes. Virology 206: 935–944. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

