Binning sequences using very sparse labels within a metagenome
- PMID: 18442374
- PMCID: PMC2383919
- DOI: 10.1186/1471-2105-9-215
Binning sequences using very sparse labels within a metagenome
Abstract
Background: In metagenomic studies, a process called binning is necessary to assign contigs that belong to multiple species to their respective phylogenetic groups. Most of the current methods of binning, such as BLAST, k-mer and PhyloPythia, involve assigning sequence fragments by comparing sequence similarity or sequence composition with already-sequenced genomes that are still far from comprehensive. We propose a semi-supervised seeding method for binning that does not depend on knowledge of completed genomes. Instead, it extracts the flanking sequences of highly conserved 16S rRNA from the metagenome and uses them as seeds (labels) to assign other reads based on their compositional similarity.
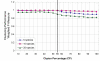
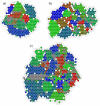
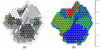
Results: The proposed seeding method is implemented on an unsupervised Growing Self-Organising Map (GSOM), and called Seeded GSOM (S-GSOM). We compared it with four well-known semi-supervised learning methods in a preliminary test, separating random-length prokaryotic sequence fragments sampled from the NCBI genome database. We identified the flanking sequences of the highly conserved 16S rRNA as suitable seeds that could be used to group the sequence fragments according to their species. S-GSOM showed superior performance compared to the semi-supervised methods tested. Additionally, S-GSOM may also be used to visually identify some species that do not have seeds. The proposed method was then applied to simulated metagenomic datasets using two different confidence threshold settings and compared with PhyloPythia, k-mer and BLAST. At the reference taxonomic level Order, S-GSOM outperformed all k-mer and BLAST results and showed comparable results with PhyloPythia for each of the corresponding confidence settings, where S-GSOM performed better than PhyloPythia in the >/= 10 reads datasets and comparable in the > or = 8 kb benchmark tests.
Conclusion: In the task of binning using semi-supervised learning methods, results indicate S-GSOM to be the best of the methods tested. Most importantly, the proposed method does not require knowledge from known genomes and uses only very few labels (one per species is sufficient in most cases), which are extracted from the metagenome itself. These advantages make it a very attractive binning method. S-GSOM outperformed the binning methods that depend on already-sequenced genomes, and compares well to the current most advanced binning method, PhyloPythia.
Figures


References
-
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers Y-H, Smith HOB. Environmental Genome Shotgun Sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. - DOI - PubMed
-
- Woyke T, Teeling H, Ivanova NN, Huntemann M, Richter M, Gloeckner FO, Boffelli D, Anderson IJ, Barry KW, Shapiro HJ, Szeto E, Kyrpides NC, Mussmann M, Amann R, Bergin C, Ruehland C, Rubin EM, Dubilier N. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature. 2006;443:950–955. doi: 10.1038/nature05192. - DOI - PubMed
-
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, Beeson K, Tran B, Smith H, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, Li K, Kravitz S, Heidelberg JF, Utterback T, Rogers Y-H, Falcón LI, Souza V, Bonilla-Rosso G, Eguiarte LE, Karl DM, Sathyendranath S, et al. The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biology. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

