Application of bioinformatics-coupled experimental analysis reveals a new transport-competent nuclear localization signal in the nucleoprotein of influenza A virus strain
- PMID: 18442378
- PMCID: PMC2386121
- DOI: 10.1186/1471-2121-9-22
Application of bioinformatics-coupled experimental analysis reveals a new transport-competent nuclear localization signal in the nucleoprotein of influenza A virus strain
Abstract
Background: Two nuclear localization sequences (NLS) in influenza A virus nucleoprotein (NP) have been demonstrated to be critical for nuclear import of NP and viral ribonucleoprotein complexes. However, a deletion mutant lacking these two signals was still able to localize to the nucleus suggesting the presence of yet another (a third) potential NLS in the NP protein. In order to identify the nature of this potential NLS signal in the NP of a WS/33L influenza virus A strain, we utilized the tools of bioinformatics coupled with functional experimental analyses in the present study.
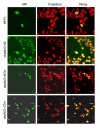
Results: Comparison of the deduced aa sequence of NP of WS/33L strain with the published WS/33 NP sequences revealed that a single amino acid (aa) change (Met to Arg) at position 105 results in converting the flanking regions (between aa position 90-121, a 32-residue stretch) into two classical overlapping bipartite NLS (obpNLS). GenBank search revealed that 9 out of 500 published NP sequences contain a similar Arg at position 105 (instead of Met) with a 100% homology to the obpNLS region. Various NP-green fluorescent protein (GFP) fusion constructs with and without the signal (obpNLS-Arg105) were utilized to understand the functional nature of this signal. We analyzed the transport competency of the expressed chimeric proteins in terms of their cellular localization by confocal immunofluorescence assay. Our analysis revealed that all NP-GFP constructs containing the wild-type (R105) sequence localized predominantly to the nucleus. Constructs lacking the obpNLS or constructs with reverse mutation (R105 to M105) on the other hand exhibited predominant cytoplasmic localization pattern. Interestingly, when the 32 aa obpNLS was fused with an unrelated viral protein (rotavirus NSP6) that has been known to be cytoplasmic protein, the chimeric protein (obpNLS-NSP6) was efficiently transported into the nucleus, indicating an efficient nuclear transport function of the 32-residue obpNLS in the NP of WS/33L strain of influenza A virus.
Conclusion: This report while not only establishing a new NLS in the influenza A virus strain, it also reinforces the idea that proper application of bioinformatics-coupled experimental analysis serves as a powerful tool in identifying new functional signals in proteins of interest.
Figures




Similar articles
-
Importin α3/Qip1 is involved in multiplication of mutant influenza virus with alanine mutation at amino acid 9 independently of nuclear transport function.PLoS One. 2013;8(1):e55765. doi: 10.1371/journal.pone.0055765. Epub 2013 Jan 30. PLoS One. 2013. PMID: 23383277 Free PMC article.
-
A classical bipartite nuclear localization signal on Thogoto and influenza A virus nucleoproteins.Virology. 1998 Oct 10;250(1):9-18. doi: 10.1006/viro.1998.9329. Virology. 1998. PMID: 9770415
-
Nuclear import and export of influenza virus nucleoprotein.J Virol. 1997 Dec;71(12):9690-700. doi: 10.1128/JVI.71.12.9690-9700.1997. J Virol. 1997. PMID: 9371635 Free PMC article.
-
Structure and sequence analysis of influenza A virus nucleoprotein.Sci China C Life Sci. 2009 May;52(5):439-49. doi: 10.1007/s11427-009-0064-x. Epub 2009 May 27. Sci China C Life Sci. 2009. PMID: 19471866 Review.
-
Antiviral drug development by targeting RNA binding site, oligomerization and nuclear export of influenza nucleoprotein.Int J Biol Macromol. 2024 Dec;282(Pt 4):136996. doi: 10.1016/j.ijbiomac.2024.136996. Epub 2024 Oct 31. Int J Biol Macromol. 2024. PMID: 39486729 Review.
Cited by
-
Eukaryotic Translation Elongation Factor 1 Delta Inhibits the Nuclear Import of the Nucleoprotein and PA-PB1 Heterodimer of Influenza A Virus.J Virol. 2020 Dec 22;95(2):e01391-20. doi: 10.1128/JVI.01391-20. Print 2020 Dec 22. J Virol. 2020. PMID: 33087462 Free PMC article.
-
A Novel Antiviral Target Structure Involved in the RNA Binding, Dimerization, and Nuclear Export Functions of the Influenza A Virus Nucleoprotein.PLoS Pathog. 2015 Jul 29;11(7):e1005062. doi: 10.1371/journal.ppat.1005062. eCollection 2015 Jul. PLoS Pathog. 2015. PMID: 26222066 Free PMC article.
-
Organelle dynamics and viral infections: at cross roads.Microbes Infect. 2019 Jan-Feb;21(1):20-32. doi: 10.1016/j.micinf.2018.06.002. Epub 2018 Jun 25. Microbes Infect. 2019. PMID: 29953921 Free PMC article. Review.
-
Importin α3/Qip1 is involved in multiplication of mutant influenza virus with alanine mutation at amino acid 9 independently of nuclear transport function.PLoS One. 2013;8(1):e55765. doi: 10.1371/journal.pone.0055765. Epub 2013 Jan 30. PLoS One. 2013. PMID: 23383277 Free PMC article.
-
New-generation screening assays for the detection of anti-influenza compounds targeting viral and host functions.Antiviral Res. 2013 Oct;100(1):120-32. doi: 10.1016/j.antiviral.2013.07.018. Epub 2013 Aug 6. Antiviral Res. 2013. PMID: 23933115 Free PMC article. Review.
References
-
- Palese P, Shaw ML. Orthomyxoviridae: The viruses and their replication. In: Knipes D, Howley P, editor. Field's Virology. 5. Vol. 2. Philadelphia: Lipincott Williamson and Wilkinson; 2007. pp. 1647–1689.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

