Potential role of miR-9 and miR-223 in recurrent ovarian cancer
- PMID: 18442408
- PMCID: PMC2383925
- DOI: 10.1186/1476-4598-7-35
Potential role of miR-9 and miR-223 in recurrent ovarian cancer
Abstract
Background: MicroRNAs (miRNAs) are small, noncoding RNAs that negatively regulate gene expression by binding to target mRNAs. miRNAs have not been comprehensively studied in recurrent ovarian cancer, yet an incurable disease.
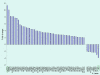
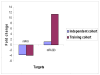
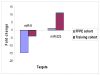
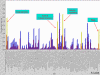
Results: Using real-time RT-PCR, we obtained distinct miRNA expression profiles between primary and recurrent serous papillary ovarian adenocarcinomas (n = 6) in a subset of samples previously used in a transcriptome approach. Expression levels of top dysregulated miRNA genes, miR-223 and miR-9, were examined using TaqMan PCR in independent cohorts of fresh frozen (n = 18) and FFPE serous ovarian tumours (n = 22). Concordance was observed on TaqMan analysis for miR-223 and miR-9 between the training cohort and the independent test cohorts. Target prediction analysis for the above miRNA "recurrent metastatic signature" identified genes previously validated in our transcriptome study. Common biological pathways well characterised in ovarian cancer were shared by miR-9 and miR-223 lists of predicted target genes. We provide strong evidence that miR-9 acts as a putative tumour suppressor gene in recurrent ovarian cancer. Components of the miRNA processing machinery, such as Dicer and Drosha are not responsible for miRNA deregulation in recurrent ovarian cancer, as deluded by TaqMan and immunohistochemistry.
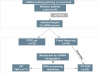
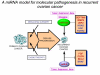
Conclusion: We propose a miRNA model for the molecular pathogenesis of recurrent ovarian cancer. Some of the differentially deregulated miRNAs identified correlate with our previous transcriptome findings. Based on integrated transcriptome and miRNA analysis, miR-9 and miR-223 can be of potential importance as biomarkers in recurrent ovarian cancer.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

