A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells
- PMID: 18442996
- PMCID: PMC2425496
- DOI: 10.1093/nar/gkn108
A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells
Abstract
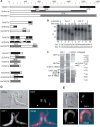
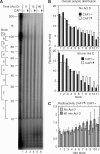
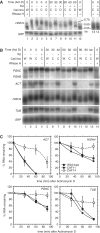
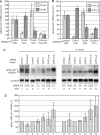
The eukaryotic Ccr4/Caf1/Not complex is involved in deadenylation of mRNAs. The Caf1 and Ccr4 subunits both potentially have deadenylating enzyme activity. We investigate here the roles of Ccr4 and Caf1 in deadenylation in two organisms that separated early in eukaryotic evolution: humans and trypanosomes. In Trypanosoma brucei, we found a complex containing CAF1, NOT1, NOT2 and NOT5, DHH1 and a possible homologue of Caf130; no homologue of Ccr4 was found. Trypanosome CAF1 has deadenylation activity, and is essential for cell survival. Depletion of trypanosome CAF1 delayed deadenylation and degradation of constitutively expressed mRNAs. Human cells have two isozymes of Caf1: simultaneous depletion of both inhibited degradation of an unstable reporter mRNA. In both species, depletion of Caf1 homologues inhibited deadenylation of bulk RNA and resulted in an increase in average poly(A) tail length.
Figures


Similar articles
-
The role of deadenylation in the degradation of unstable mRNAs in trypanosomes.Nucleic Acids Res. 2009 Sep;37(16):5511-28. doi: 10.1093/nar/gkp571. Epub 2009 Jul 13. Nucleic Acids Res. 2009. PMID: 19596809 Free PMC article.
-
The roles of 3'-exoribonucleases and the exosome in trypanosome mRNA degradation.RNA. 2013 Jul;19(7):937-47. doi: 10.1261/rna.038430.113. Epub 2013 May 22. RNA. 2013. PMID: 23697549 Free PMC article.
-
CAF1 plays an important role in mRNA deadenylation separate from its contact to CCR4.Nucleic Acids Res. 2007;35(9):3002-15. doi: 10.1093/nar/gkm196. Epub 2007 Apr 16. Nucleic Acids Res. 2007. PMID: 17439972 Free PMC article.
-
The CAF1-NOT complex of trypanosomes.Front Genet. 2014 Jan 2;4:299. doi: 10.3389/fgene.2013.00299. Front Genet. 2014. PMID: 24427169 Free PMC article. Review.
-
Mechanisms of deadenylation-dependent decay.Wiley Interdiscip Rev RNA. 2011 Mar-Apr;2(2):167-83. doi: 10.1002/wrna.40. Epub 2010 Sep 15. Wiley Interdiscip Rev RNA. 2011. PMID: 21957004 Free PMC article. Review.
Cited by
-
Identification of the HSP70-II gene in Leishmania braziliensis HSP70 locus: genomic organization and UTRs characterization.Parasit Vectors. 2011 Aug 26;4:166. doi: 10.1186/1756-3305-4-166. Parasit Vectors. 2011. PMID: 21871099 Free PMC article.
-
Ciphers and Executioners: How 3'-Untranslated Regions Determine the Fate of Messenger RNAs.Front Genet. 2019 Jan 24;10:6. doi: 10.3389/fgene.2019.00006. eCollection 2019. Front Genet. 2019. PMID: 30740123 Free PMC article. Review.
-
Genome-wide RIP-Chip analysis of translational repressor-bound mRNAs in the Plasmodium gametocyte.Genome Biol. 2014 Nov 3;15(11):493. doi: 10.1186/s13059-014-0493-0. Genome Biol. 2014. PMID: 25418785 Free PMC article.
-
Dynamic Control of Dendritic mRNA Expression by CNOT7 Regulates Synaptic Efficacy and Higher Cognitive Function.Cell Rep. 2017 Jul 18;20(3):683-696. doi: 10.1016/j.celrep.2017.06.078. Cell Rep. 2017. PMID: 28723570 Free PMC article.
-
Heat shock causes a decrease in polysomes and the appearance of stress granules in trypanosomes independently of eIF2(alpha) phosphorylation at Thr169.J Cell Sci. 2008 Sep 15;121(Pt 18):3002-14. doi: 10.1242/jcs.031823. Epub 2008 Aug 19. J Cell Sci. 2008. PMID: 18713834 Free PMC article.
References
-
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. - PubMed
-
- Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

