An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors
- PMID: 18443585
- PMCID: PMC2912221
- DOI: 10.1038/ng.127
An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors
Abstract
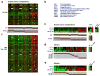
Cancer cells possess traits reminiscent of those ascribed to normal stem cells. It is unclear, however, whether these phenotypic similarities reflect the activity of common molecular pathways. Here, we analyze the enrichment patterns of gene sets associated with embryonic stem (ES) cell identity in the expression profiles of various human tumor types. We find that histologically poorly differentiated tumors show preferential overexpression of genes normally enriched in ES cells, combined with preferential repression of Polycomb-regulated genes. Moreover, activation targets of Nanog, Oct4, Sox2 and c-Myc are more frequently overexpressed in poorly differentiated tumors than in well-differentiated tumors. In breast cancers, this ES-like signature is associated with high-grade estrogen receptor (ER)-negative tumors, often of the basal-like subtype, and with poor clinical outcome. The ES signature is also present in poorly differentiated glioblastomas and bladder carcinomas. We identify a subset of ES cell-associated transcription regulators that are highly expressed in poorly differentiated tumors. Our results reveal a previously unknown link between genes associated with ES cell identity and the histopathological traits of tumors and support the possibility that these genes contribute to stem cell-like phenotypes shown by many tumors.
Figures




References
-
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. - PubMed
-
- Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432:324–331. - PubMed
-
- Stingl J, Caldas C. Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. Nat Rev Cancer. 2007;7:791–799. - PubMed
-
- Lobo NA, Shimono Y, Qian D, Clarke MF. The Biology of Cancer Stem Cells. Annu Rev Cell Dev Biol. 2007 - PubMed
-
- Andres AC, et al. Ha-ras and c-myc oncogene expression interferes with morphological and functional differentiation of mammary epithelial cells in single and double transgenic mice. Genes Dev. 1988;2:1486–1495. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

