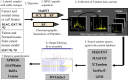
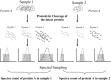
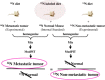
Cancer proteomics by quantitative shotgun proteomics
- PMID: 18443658
- PMCID: PMC2352161
- DOI: 10.1016/j.molonc.2007.05.001
Cancer proteomics by quantitative shotgun proteomics
Abstract
A major scientific challenge at the present time for cancer research is the determination of the underlying biological basis for cancer development. It is further complicated by the heterogeneity of cancer's origin. Understanding the molecular basis of cancer requires studying the dynamic and spatial interactions among proteins in cells, signaling events among cancer cells, and interactions between the cancer cells and the tumor microenvironment. Recently, it has been proposed that large-scale protein expression analysis of cancer cell proteomes promises to be valuable for investigating mechanisms of cancer transformation. Advances in mass spectrometry technologies and bioinformatics tools provide a tremendous opportunity to qualitatively and quantitatively interrogate dynamic protein-protein interactions and differential regulation of cellular signaling pathways associated with tumor development. In this review, progress in shotgun proteomics technologies for examining the molecular basis of cancer development will be presented and discussed.
Keywords: cancer cells; mass spectrometry; protein profiling; quantitative proteomics; shotgun proteomics.
Figures
Similar articles
-
Recent progress in mass spectrometry proteomics for biomedical research.Sci China Life Sci. 2017 Oct;60(10):1093-1113. doi: 10.1007/s11427-017-9175-2. Epub 2017 Oct 13. Sci China Life Sci. 2017. PMID: 29039124 Review.
-
Application of targeted mass spectrometry in bottom-up proteomics for systems biology research.J Proteomics. 2018 Oct 30;189:75-90. doi: 10.1016/j.jprot.2018.02.008. Epub 2018 Feb 13. J Proteomics. 2018. PMID: 29452276 Free PMC article. Review.
-
Shotgun Proteomics and Mass Spectrometry as a Tool for Protein Identification and Profiling of Bio-Carrier-Based Therapeutics on Human Cancer Cells.Methods Mol Biol. 2021;2211:233-240. doi: 10.1007/978-1-0716-0943-9_16. Methods Mol Biol. 2021. PMID: 33336281
-
Emerging Proteomic Technologies Provide Enormous and Underutilized Potential for Brain Cancer Research.Mol Cell Proteomics. 2016 Feb;15(2):362-7. doi: 10.1074/mcp.R115.053884. Epub 2015 Sep 25. Mol Cell Proteomics. 2016. PMID: 26407994 Free PMC article. Review.
-
A review of proteomics in cancer research.Asian Pac J Cancer Prev. 2005 Apr-Jun;6(2):113-7. Asian Pac J Cancer Prev. 2005. PMID: 16101316 Review.
Cited by
-
Application of Proteomics in Pancreatic Ductal Adenocarcinoma Biomarker Investigations: A Review.Int J Mol Sci. 2022 Feb 14;23(4):2093. doi: 10.3390/ijms23042093. Int J Mol Sci. 2022. PMID: 35216204 Free PMC article. Review.
-
The effects of nonignorable missing data on label-free mass spectrometry proteomics experiments.Ann Appl Stat. 2018 Dec;12(4):2075-2095. doi: 10.1214/18-AOAS1144. Epub 2018 Nov 13. Ann Appl Stat. 2018. PMID: 30473739 Free PMC article.
-
A single lysis solution for the analysis of tissue samples by different proteomic technologies.Mol Oncol. 2008 Dec;2(4):368-79. doi: 10.1016/j.molonc.2008.09.003. Epub 2008 Oct 2. Mol Oncol. 2008. PMID: 19383358 Free PMC article.
-
Targeted proteomic strategy for clinical biomarker discovery.Mol Oncol. 2009 Feb;3(1):33-44. doi: 10.1016/j.molonc.2008.12.001. Epub 2008 Dec 11. Mol Oncol. 2009. PMID: 19383365 Free PMC article. Review.
-
Refinements of LC-MS/MS Spectral Counting Statistics Improve Quantification of Low Abundance Proteins.Sci Rep. 2019 Sep 20;9(1):13653. doi: 10.1038/s41598-019-49665-1. Sci Rep. 2019. PMID: 31541118 Free PMC article.
References
-
- Abe, Y. , Chaen, T. , Jin, X.R. , Hamasaki, T. , Hamasaki, N. , 2004. Mass spectrometric analyses of transmembrane proteins in human erythrocyte membrane. J. Biochem. (Tokyo). 136, 97–106. - PubMed
-
- Aggarwal, K. , Choe, L.H. , Lee, K.H. , 2006. Shotgun proteomics using the iTRAQ isobaric tags. Brief Funct. Genomic Proteomic. 5, 112–120. - PubMed
-
- Albertson, D.G. , Collins, C. , McCormick, F. , Gray, J.W. , 2003. Chromosome aberrations in solid tumors. Nat. Genet.. 34, 369–376. - PubMed
-
- Amanchy, R. , Kalume, D.E. , Iwahori, A. , Zhong, J. , Pandey, A. , 2005. Phosphoproteome analysis of HeLa cells using stable isotope labeling with amino acids in cell culture (SILAC). J. Proteome Res.. 4, 1661–1671. - PubMed
-
- Anderson, L. , Hunter, C.L. , 2006. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteomics. 5, 573–588. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources