Evaluation of biological pathways involved in chemotherapy response in breast cancer
- PMID: 18445275
- PMCID: PMC2397539
- DOI: 10.1186/bcr2088
Evaluation of biological pathways involved in chemotherapy response in breast cancer
Abstract
Introduction: Our goal was to examine the association between biological pathways and response to chemotherapy in estrogen receptor-positive (ER+) and ER-negative (ER-) breast tumors separately.
Methods: Gene set enrichment analysis including 852 predefined gene sets was applied to gene expression data from 51 ER- and 82 ER+ breast tumors that were all treated with a preoperative paclitaxel, 5-fluoruracil, doxorubicin, and cyclophosphamide chemotherapy.
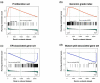
Results: Twenty-seven (53%) ER- and 7 (9%) ER+ patients had pathologic complete response (pCR) to therapy. Among the ER- tumors, a proliferation gene signature (false discovery rate [FDR] q = 0.1), the genomic grade index (FDR q = 0.044), and the E2F3 pathway signature (FDR q = 0.22, P = 0.07) were enriched in the pCR group. Among the ER+ tumors, the proliferation signature (FDR q = 0.001) and the genomic grade index (FDR q = 0.015) were also significantly enriched in cases with pCR. Ki67 expression, as single gene marker of proliferation, did not provide the same information as the entire proliferation signature. An ER-associated gene set (FDR q = 0.03) and a mutant p53 gene signature (FDR q = 0.0019) were enriched in ER+ tumors with residual cancer.
Conclusion: Proliferation- and genomic grade-related gene signatures are associated with chemotherapy sensitivity in both ER- and ER+ breast tumors. Genes involved in the E2F3 pathway are associated with chemotherapy sensitivity among ER- tumors. The mutant p53 signature and expression of ER-related genes were associated with lower sensitivity to chemotherapy in ER+ breast tumors only.
Figures


References
-
- Kaufmann M, von Minckwitz G, Smith R, Valero V, Gianni L, Eiermann W, Howell A, Costa SD, Beuzeboc P, Untch M, Blohmer JU, Sinn HP, Sittek R, Souchon R, Tulusan AH, Volm T, Senn HJ. International expert panel on the use of primary (preoperative) systemic treatment of operable breast cancer: review and recommendations. J Clin Onc. 2003;21:2600–2608. doi: 10.1200/JCO.2003.01.136. - DOI - PubMed
-
- Mazouni C, Kau SW, Frye D, Andre F, Buchholz TA, Symmans WF, Anderson K, Hess KR, Gonzalez-Angulo AM, Hortobagyi GN, Buzdar AU, Pusztai L. Inclusion of taxanes, particularly weekly paclitaxel, in preoperative chemotherapy improves pathologic complete response rate in ER-positive breast cancers. Ann Oncol. 2007;18:878–880. doi: 10.1093/annonc/mdm008. - DOI - PubMed
-
- Kuerer HM, Newman LA, Smith TL, Ames FC, Hunt KK, Dhingra K, Theriault RL, Singh G, Binkley SM, Sneige N, Buchholz TA, Ross MI, McNeese MD, Buzdar AU, Hortobagyi GN, Singletary SE. Clinical course of breast cancer patients with complete pathologic primary tumor and axillary lymph node response to doxorubicin-based neoadjuvant chemotherapy. J Clin Oncol. 1999;17:460–469. - PubMed
-
- Hess KR, Anderson K, Symmans WF, Valero V, Ibrahim N, Mejia JA, Booser D, Theriault RL, Buzdar AU, Dempsey PJ, Rouzier R, Sneige N, Ross JS, Vidaurre T, Gómez HL, Hortobagyi GN, Pusztai L. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24:4236–4244. doi: 10.1200/JCO.2006.05.6861. - DOI - PubMed
-
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

