In vivo consequences of deleting EGF repeats 8-12 including the ligand binding domain of mouse Notch1
- PMID: 18445292
- PMCID: PMC2390518
- DOI: 10.1186/1471-213X-8-48
In vivo consequences of deleting EGF repeats 8-12 including the ligand binding domain of mouse Notch1
Abstract
Background: Notch signaling is highly conserved in the metazoa and is critical for many cell fate decisions. Notch activation occurs following ligand binding to Notch extracellular domain. In vitro binding assays have identified epidermal growth factor (EGF) repeats 11 and 12 as the ligand binding domain of Drosophila Notch. Here we show that an internal deletion in mouse Notch1 of EGF repeats 8-12, including the putative ligand binding domain (lbd), is an inactivating mutation in vivo. We also show that maternal and zygotic Notch1(lbd/lbd) mutant embryos develop through gastrulation to mid-gestation.
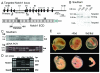
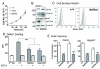
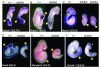
Results: Notch1(lbd/lbd) embryos died at mid-gestation with a phenotype indistinguishable from Notch1 null mutants. In embryonic stem (ES) cells, Notch1(lbd) was expressed on the cell surface at levels equivalent to wild type Notch1, but Delta1 binding was reduced to the same level as in Notch1 null cells. In an ES cell co-culture assay, Notch signaling induced by Jagged1 or Delta1 was reduced to a similar level in Notch1(lbd) and Notch1 null cells. However, the Notch1(lbd/lbd) allele was expressed similarly to wild type Notch1 in Notch1(lbd/lbd) ES cells and embryos at E8.75, indicating that Notch1 signaling is not essential for the Notch1 gene to be expressed. In addition, maternal and zygotic Notch1 mutant blastocysts developed through gastrulation.
Conclusion: Mouse Notch1 lacking the ligand binding domain is expressed at the cell surface but does not signal in response to the canonical Notch ligands Delta1 and Jagged1. Homozygous Notch1(lbd/lbd) mutant embryos die at approximately E10 similar to Notch1 null embryos. While Notch1 is expressed in oocytes and blastocysts, Notch1 signaling via canonical ligands is dispensable during oogenesis, blastogenesis, implantation and gastrulation.
Figures

References
-
- Lai EC. Notch signaling: control of cell communication and cell fate. Development (Cambridge, England) 2004;131:965–973. - PubMed
-
- Schweisguth F. Regulation of notch signaling activity. Curr Biol. 2004;14:R129–38. - PubMed
-
- Xu A, Lei L, Irvine KD. Regions of Drosophila Notch That Contribute to Ligand Binding and the Modulatory Influence of Fringe. 2005;280:30158–30165. - PubMed
-
- Hartmann D, de Strooper B, Serneels L, Craessaerts K, Herreman A, Annaert W, Umans L, Lubke T, Lena Illert A, von Figura K, Saftig P. The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum Mol Genet. 2002;11:2615–2624. doi: 10.1093/hmg/11.21.2615. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

