Design of protein function leaps by directed domain interface evolution
- PMID: 18445649
- PMCID: PMC2373342
- DOI: 10.1073/pnas.0801097105
Design of protein function leaps by directed domain interface evolution
Abstract
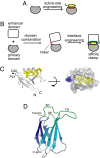
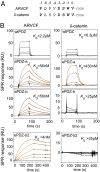
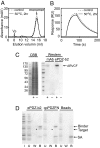
Most natural proteins performing sophisticated tasks contain multiple domains where an active site is located at the domain interface. Comparative structural analyses suggest that major leaps in protein function occur through gene recombination events that connect two or more protein domains to generate a new active site, frequently occurring at the newly created domain interface. However, such functional leaps by combination of unrelated domains have not been directly demonstrated. Here we show that highly specific and complex protein functions can be generated by joining a low-affinity peptide-binding domain with a functionally inert second domain and subsequently optimizing the domain interface. These directed evolution processes dramatically enhanced both affinity and specificity to a level unattainable with a single domain, corresponding to >500-fold and >2,000-fold increases of affinity and specificity, respectively. An x-ray crystal structure revealed that the resulting "affinity clamp" had clamshell architecture as designed, with large additional binding surface contributed by the second domain. The affinity clamps having a single-nanomolar dissociation constant outperformed a monoclonal antibody in immunochemical applications. This work establishes evolutionary paths from isolated domains with primitive function to multidomain proteins with sophisticated function and introduces a new protein-engineering concept that allows for the generation of highly functional affinity reagents to a predefined target. The prevalence and variety of natural interaction domains suggest that numerous new functions can be designed by using directed domain interface evolution.
Conflict of interest statement
Conflict of interest statement: J.H., A.K., and S.K. are named as the inventors of a patent application on the technology described in this article.
Figures

References
-
- Todd AE, Orengo CA, Thornton JM. Sequence and structural differences between enzyme and nonenzyme homologs. Structure. 2002;10:1435–1451. - PubMed
-
- Pluckthun A, Mayo SL. The design of evolution and the evolution of design. Curr Opin Struct Biol. 2007;17:451–453.
-
- Kortemme T, Baker D. Computational design of protein-protein interactions. Curr Opin Chem Biol. 2004;8:91–97. - PubMed
-
- Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. Making antibodies by phage display technology. Annu Rev Immunol. 1994;12:433–455. - PubMed
-
- Binz HK, Amstutz P, Pluckthun A. Engineering novel binding proteins from nonimmunoglobulin domains. Nat Biotechnol. 2005;23:1257–1268. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

