Phenotypic transcription factors epigenetically mediate cell growth control
- PMID: 18445650
- PMCID: PMC2373311
- DOI: 10.1073/pnas.0800970105
Phenotypic transcription factors epigenetically mediate cell growth control
Abstract
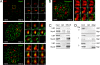
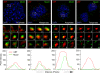
Ribosomal RNA (rRNA) genes are down-regulated during osteogenesis, myogenesis, and adipogenesis, necessitating a mechanistic understanding of interrelationships between growth control and phenotype commitment. Here, we show that cell fate-determining factors [MyoD, myogenin (Mgn), Runx2, C/EBPbeta] occupy rDNA loci and suppress rRNA expression during lineage progression, concomitant with decreased rRNA expression and reciprocal loss of occupancy by c-Myc, a proliferation-specific activator of rRNA transcription. We find interaction of phenotypic factors with the polymerase I activator upstream binding factor UBF-1 at interphase nucleoli, and this interaction is epigenetically retained on mitotic chromosomes at nucleolar organizing regions. Ectopic expression and RNA interference establish that MyoD, Mgn, Runx2, and C/EBPbeta each functionally suppress rRNA genes and global protein synthesis. We conclude that epigenetic control of ribosomal biogenesis by lineage-specific differentiation factors is a general developmental mechanism for coordinate control of cell growth and phenotype.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



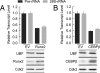
References
-
- Hernandez-Verdun D, Roussel P. Regulators of nucleolar functions. Prog Cell Cycle Res. 2003;5:301–308. - PubMed
-
- Lee MH, et al. Transient up-regulation of CBFA1 in response to bone morphogenetic protein-2 and transforming growth factor β1 in C2C12 myogenic cells coincides with suppression of the myogenic phenotype but is not sufficient for osteoblast differentiation. J Cell Biochem. 1999;73:114–125. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

