Secondary chromosomal attachment site and tandem integration of the mobilizable Salmonella genomic island 1
- PMID: 18446190
- PMCID: PMC2297512
- DOI: 10.1371/journal.pone.0002060
Secondary chromosomal attachment site and tandem integration of the mobilizable Salmonella genomic island 1
Abstract
Background: The Salmonella genomic island 1 is an integrative mobilizable element (IME) originally identified in epidemic multidrug-resistant Salmonella enterica serovar Typhimurium (S. Typhimurium) DT104. SGI1 contains a complex integron, which confers various multidrug resistance phenotypes due to its genetic plasticity. Previous studies have shown that SGI1 integrates site-specifically into the S. enterica, Escherichia coli, or Proteus mirabilis chromosome at the 3' end of thdF gene (attB site).
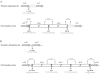
Methodology/principal findings: Here, we report the transfer of SGI1 to a Delta thdF mutant of S. Typhimurium LT2. In the absence of thdF, the frequency of transconjugant formation was reduced by around thirty times of magnitude. Through DNA sequencing SGI1 was shown to integrate specifically into a secondary attachment site (2(nd)attB), which is located in the intergenic region between the chromosomal sodB and purR genes. At this 2(nd)attB site, we found that a significant fraction of SGI1 transconjugants (43% of wild type and 100% of Delta thdF mutant) contained tandem SGI1 arrays. Moreover, in wild type S. Typhimurium LT2 transconjugants, SGI1 integrated into both attachment sites, i.e., thdF and sodB-purR. The formation of SGI1 tandem arrays occurred in both specific attB sites. There was heterogeneity in the size of the SGI1 tandem arrays detected in single transconjugant colonies. Some arrays consisted as far as six SGI1s arranged in tandem. These tandem arrays were shown to persist during serial passages with or without antibiotic selection pressure.
Conclusions/significance: The ability of integration into two distinct chromosomal sites and tandem array formation of SGI1 could contribute to its spread and persistence. The existence of a secondary attachment site in the Salmonella chromosome has potential implications for the mobility of SGI1, which may integrate in other attachment sites of other bacterial pathogens that do not possess the 1(st) or 2(nd) specific SGI1 attB sites of Salmonella.
Conflict of interest statement
Figures


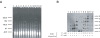
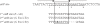
References
-
- Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol. 2004;2:414–424. - PubMed
-
- Burrus V, Pavlovic G, Decaris B, Guédon G. Conjugative transposons: the tip of the iceberg. Mol Microbiol. 2002;46:601–610. - PubMed
-
- Burrus V, Waldor MK. Shaping bacterial genomes with integrative and conjugative elements Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol. 2004;155:376–386. - PubMed
-
- Boyd D, Peters GA, Cloeckaert A, Sidi Boumedine K, Chaslus-Dancla E, et al. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J Bacteriol. 2001;183:5725–5732. - PMC - PubMed
-
- Mulvey MR, Boyd DA, Olson AB, Doublet B, Cloeckaert A. The genetics of Salmonella genomic island 1. Microbes Infect. 2006;8:1915–1922. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

