The Nod-like receptor (NLR) family: a tale of similarities and differences
- PMID: 18446235
- PMCID: PMC2323615
- DOI: 10.1371/journal.pone.0002119
The Nod-like receptor (NLR) family: a tale of similarities and differences
Abstract
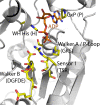
Innate immunity represents an important system with a variety of vital processes at the core of many diseases. In recent years, the central role of the Nod-like receptor (NLR) protein family became increasingly appreciated in innate immune responses. NLRs are classified as part of the signal transduction ATPases with numerous domains (STAND) clade within the AAA+ ATPase family. They typically feature an N-terminal effector domain, a central nucleotide-binding domain (NACHT) and a C-terminal ligand-binding region that is composed of several leucine-rich repeats (LRRs). NLRs are believed to initiate or regulate host defense pathways through formation of signaling platforms that subsequently trigger the activation of inflammatory caspases and NF-kB. Despite their fundamental role in orchestrating key pathways in innate immunity, their mode of action in molecular terms remains largely unknown. Here we present the first comprehensive sequence and structure modeling analysis of NLR proteins, revealing that NLRs possess a domain architecture similar to the apoptotic initiator protein Apaf-1. Apaf-1 performs its cellular function by the formation of a heptameric platform, dubbed apoptosome, ultimately triggering the controlled demise of the affected cell. The mechanism of apoptosome formation by Apaf-1 potentially offers insight into the activation mechanisms of NLR proteins. Multiple sequence alignment analysis and homology modeling revealed Apaf-1-like structural features in most members of the NLR family, suggesting a similar biochemical behaviour in catalytic activity and oligomerization. Evolutionary tree comparisons substantiate the conservation of characteristic functional regions within the NLR family and are in good agreement with domain distributions found in distinct NLRs. Importantly, the analysis of LRR domains reveals surprisingly low conservation levels among putative ligand-binding motifs. The same is true for the effector domains exhibiting distinct interfaces ensuring specific interactions with downstream target proteins. All together these factors suggest specific biological functions for individual NLRs.
Conflict of interest statement
Figures

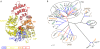
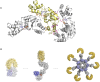


References
-
- Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. - PubMed
-
- Fritz JH, Ferrero RL, Philpott DJ, Girardin SE. Nod-like proteins in immunity, inflammation and disease. Nat Immunol. 2006;7:1250–1257. - PubMed
-
- Werts C, Girardin SE, Philpott DJ. TIR, CARD and PYRIN: three domains for an antimicrobial triad. Cell Death Differ. 2006;13:798–815. - PubMed
-
- Inohara N, Nunez G. NODs: intracellular proteins involved in inflammation and apoptosis. Nat Rev Immunol. 2003;3:371–382. - PubMed
-
- Mariathasan S, Newton K, Monack DM, Vucic D, French DM, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

