A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay
- PMID: 18447585
- PMCID: PMC2689706
- DOI: 10.1371/journal.pbio.0060111
A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay
Abstract
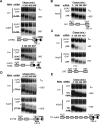
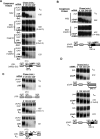
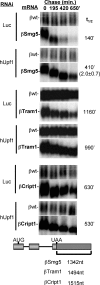
The nonsense-mediated decay (NMD) pathway subjects mRNAs with premature termination codons (PTCs) to rapid decay. The conserved Upf1-3 complex interacts with the eukaryotic translation release factors, eRF3 and eRF1, and triggers NMD when translation termination takes place at a PTC. Contrasting models postulate central roles in PTC-recognition for the exon junction complex in mammals versus the cytoplasmic poly(A)-binding protein (PABP) in other eukaryotes. Here we present evidence for a unified model for NMD, in which PTC recognition in human cells is mediated by a competition between 3' UTR-associated factors that stimulate or antagonize recruitment of the Upf complex to the terminating ribosome. We identify cytoplasmic PABP as a human NMD antagonizing factor, which inhibits the interaction between eRF3 and Upf1 in vitro and prevents NMD in cells when positioned in proximity to the termination codon. Surprisingly, only when an extended 3' UTR places cytoplasmic PABP distally to the termination codon does a downstream exon junction complex enhance NMD, likely through increasing the affinity of Upf proteins for the 3' UTR. Interestingly, while an artificial 3' UTR of >420 nucleotides triggers NMD, a large subset of human mRNAs contain longer 3' UTRs but evade NMD. We speculate that these have evolved to concentrate NMD-inhibiting factors, such as PABP, in spatial proximity of the termination codon.
Conflict of interest statement
Figures



References
-
- Amrani N, Sachs MS, Jacobson A. Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol. 2006;7:415–425. - PubMed
-
- Conti E, Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr Opin Cell Biol. 2005;17:316–325. - PubMed
-
- Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. - PubMed
-
- Sharifi NA, Dietz HC. Physiologic substrates and functions for mammalian NMD. In: Maquat LE, editor. Nonsense-mediated mRNA decay. Georgetown, Texas: Landes Bioscience; 2006. pp. 97–106.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

