The identification of potential factors associated with the development of type 2 diabetes: a quantitative proteomics approach
- PMID: 18448419
- PMCID: PMC2500228
- DOI: 10.1074/mcp.M700478-MCP200
The identification of potential factors associated with the development of type 2 diabetes: a quantitative proteomics approach
Abstract
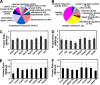
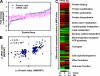
Type 2 diabetes (T2D) arises when pancreatic beta-cells fail to compensate for systemic insulin resistance with appropriate insulin secretion. However, the link between insulin resistance and beta-cell failure in T2D is not fully understood. To explore this association, we studied transgenic MKR mice that initially develop insulin resistance in skeletal muscle but by 8 weeks of age have T2D. In the present study, global islet protein and gene expression changes were characterized in diabetic MKR versus non-diabetic control mice at 10 weeks of age. Using a quantitative proteomics approach (isobaric tags for relative and absolute quantification (iTRAQ)), 159 proteins were differentially expressed in MKR compared with control islets. Marked up-regulation of protein biosynthesis and endoplasmic reticulum stress pathways and parallel down-regulation in insulin processing/secretion, energy utilization, and metabolism were observed. A fraction of the differentially expressed proteins identified (including GLUT2, DNAJC3, VAMP2, RAB3A, and PC1/3) were linked previously to insulin-secretory defects and T2D. However, many proteins for the first time were associated with islet dysfunction, including the unfolded protein response proteins (ERP72, ERP44, ERP29, PPIB, FKBP2, FKBP11, and DNAJB11), endoplasmic reticulum-associated degradation proteins (VCP and UFM1), and multiple proteins associated with mitochondrial energy metabolism (NDUFA9, UQCRH, COX2, COX4I1, COX5A, ATP6V1B2, ATP6V1H, ANT1, ANT2, ETFA, and ETFB). The mRNA expression level corresponding to these proteins was examined by microarray, and then a small subset was validated using quantitative real time PCR and Western blot analyses. Importantly approximately 54% of differentially expressed proteins in MKR islets (including proteins involved in proinsulin processing, protein biosynthesis, and mitochondrial oxidation) showed changes in the proteome but not transcriptome, suggesting post-transcriptional regulation. These results underscore the importance of integrated mRNA and protein expression measurements and validate the use of the iTRAQ method combined with microarray to assess global protein and gene changes involved in the development of T2D.
Figures



Similar articles
-
Characterization of Signaling Pathways Associated with Pancreatic β-cell Adaptive Flexibility in Compensation of Obesity-linked Diabetes in db/db Mice.Mol Cell Proteomics. 2020 Jun;19(6):971-993. doi: 10.1074/mcp.RA119.001882. Epub 2020 Apr 7. Mol Cell Proteomics. 2020. PMID: 32265294 Free PMC article.
-
Sex differences in islet stress responses support female β cell resilience.Mol Metab. 2023 Mar;69:101678. doi: 10.1016/j.molmet.2023.101678. Epub 2023 Jan 20. Mol Metab. 2023. PMID: 36690328 Free PMC article.
-
Molecular and metabolic evidence for mitochondrial defects associated with beta-cell dysfunction in a mouse model of type 2 diabetes.Diabetes. 2010 Feb;59(2):448-59. doi: 10.2337/db09-0129. Epub 2009 Nov 10. Diabetes. 2010. PMID: 19903739 Free PMC article.
-
Diabetes-induced Proteome Changes Throughout Development.Endocr Metab Immune Disord Drug Targets. 2019;19(6):732-743. doi: 10.2174/1871530319666190305153810. Endocr Metab Immune Disord Drug Targets. 2019. PMID: 31038056 Review.
-
MicroRNAs in islet hormone secretion.Diabetes Obes Metab. 2018 Sep;20 Suppl 2:11-19. doi: 10.1111/dom.13382. Diabetes Obes Metab. 2018. PMID: 30230181 Review.
Cited by
-
Biosynthesis, structure, and folding of the insulin precursor protein.Diabetes Obes Metab. 2018 Sep;20 Suppl 2(Suppl 2):28-50. doi: 10.1111/dom.13378. Diabetes Obes Metab. 2018. PMID: 30230185 Free PMC article. Review.
-
Endoplasmic reticulum protein 29 protects cortical neurons from apoptosis and promoting corticospinal tract regeneration to improve neural behavior via caspase and Erk signal in rats with spinal cord transection.Mol Neurobiol. 2014 Dec;50(3):1035-48. doi: 10.1007/s12035-014-8681-1. Epub 2014 May 3. Mol Neurobiol. 2014. PMID: 24794144
-
Overcoming β-Cell Dysfunction in Type 2 Diabetes Mellitus: CD36 Inhibition and Antioxidant System.Diabetes Metab J. 2025 Jan;49(1):1-12. doi: 10.4093/dmj.2024.0796. Epub 2025 Jan 1. Diabetes Metab J. 2025. PMID: 39828973 Free PMC article. Review.
-
Identification of potential biomarkers for predicting the early onset of diabetic cardiomyopathy in a mouse model.Sci Rep. 2020 Jul 23;10(1):12352. doi: 10.1038/s41598-020-69254-x. Sci Rep. 2020. PMID: 32703998 Free PMC article.
-
The pancreatic beta cell surface proteome.Diabetologia. 2012 Jul;55(7):1877-89. doi: 10.1007/s00125-012-2531-3. Epub 2012 Mar 31. Diabetologia. 2012. PMID: 22460761 Free PMC article. Review.
References
-
- Tiffin, N., Adie, E., Turner, F., Brunner, H. G., van Driel, M. A., Oti, M., Lopez-Bigas, N., Ouzounis, C., Perez-Iratxeta, C., Andrade-Navarro, M. A., Adeyemo, A., Patti, M. E., Semple, C. A., and Hide, W. ( 2006) Computational disease gene identification: a concert of methods prioritizes type 2 diabetes and obesity candidate genes. Nucleic Acids Res. 34, 3067–3081 - PMC - PubMed
-
- Zimmet, P., Alberti, K. G., and Shaw, J. ( 2001) Global and societal implications of the diabetes epidemic. Nature 414, 782–787 - PubMed
-
- De Fronzo, R. A. ( 1997) Pathogenesis of type 2 diabetes: metabolic and molecular implications for identifying diabetes genes. Diabetes Rev. 5, 177–285
-
- Eriksson, J., Franssila-Kallunki, A., Ekstrand, A., Saloranta, C., Widen, E., Schalin, C., and Groop, L. ( 1989) Early metabolic defects in persons at increased risk for non-insulin-dependent diabetes mellitus. N. Engl. J. Med. 321, 337–343 - PubMed
-
- Kahn, C. R., Vicent, D., and Doria, A. ( 1996) Genetics of non-insulin-dependent (type-II) diabetes mellitus. Annu. Rev. Med. 47, 509–531 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

