A conserved proliferating cell nuclear antigen-interacting protein sequence in Chk1 is required for checkpoint function
- PMID: 18448427
- PMCID: PMC2427339
- DOI: 10.1074/jbc.M800369200
A conserved proliferating cell nuclear antigen-interacting protein sequence in Chk1 is required for checkpoint function
Abstract
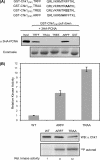
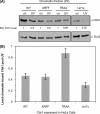
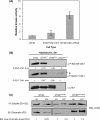
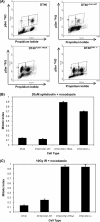
Human checkpoint kinase 1 (Chk1) is an essential kinase required for cell cycle checkpoints and for coordination of DNA synthesis. To gain insight into the mechanisms by which Chk1 carries out these functions, we used mass spectrometry to identify previously uncharacterized interacting partners of Chk1. We describe a novel interaction between Chk1 and proliferating cell nuclear antigen (PCNA), an essential component of the replication machinery. Binding between Chk1 and PCNA was reduced in the presence of hydroxyurea, suggesting that the interaction is regulated by replication stress. A highly conserved PCNA-interacting protein (PIP) box motif was identified in Chk1. The intact PIP box is required for efficient DNA damage-induced phosphorylation and release of activated Chk1 from chromatin. We find that the PIP box of Chk1 is crucial for Chk1-mediated S-M and G(2)-M checkpoint responses. In addition, we show that mutations in the PIP box of Chk1 lead to decreased rates of replication fork progression and increased aberrant replication. These findings suggest an additional mechanism by which essential components of the DNA replication machinery interact with the replication checkpoint apparatus.
Figures


References
-
- Bartek, J., and Lukas, J. (2007) Curr. Opin. Cell Biol. 19 238-245 - PubMed
-
- Capasso, H., Palermo, C., Wan, S., Rao, H., John, U. P., O'Connell, M. J., and Walworth, N. C. (2002) J. Cell Sci. 115 4555-4564 - PubMed
-
- Chen, Z., Xiao, Z., Chen, J., Ng, S. C., Sowin, T., Sham, H., Rosenberg, S., Fesik, S., and Zhang, H. (2003) Mol. Cancer Ther. 2 543-548 - PubMed
-
- Sorensen, C. S., Syljuasen, R. G., Falck, J., Schroeder, T., Ronnstrand, L., Khanna, K. K., Zhou, B. B., Bartek, J., and Lukas, J. (2003) Cancer Cell 3 247-258 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

