NOBAI: a web server for character coding of geometrical and statistical features in RNA structure
- PMID: 18448469
- PMCID: PMC2447726
- DOI: 10.1093/nar/gkn220
NOBAI: a web server for character coding of geometrical and statistical features in RNA structure
Abstract
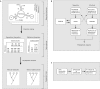
The Numeration of Objects in Biology: Alignment Inferences (NOBAI) web server provides a web interface to the applications in the NOBAI software package. This software codes topological and thermodynamic information related to the secondary structure of RNA molecules as multi-state phylogenetic characters, builds character matrices directly in NEXUS format and provides sequence randomization options. The web server is an effective tool that facilitates the search for evolutionary history embedded in the structure of functional RNA molecules. The NOBAI web server is accessible at 'http://www.manet.uiuc.edu/nobai/nobai.php'. This web site is free and open to all users and there is no login requirement.
Figures

Similar articles
-
RNA-TVcurve: a Web server for RNA secondary structure comparison based on a multi-scale similarity of its triple vector curve representation.BMC Bioinformatics. 2017 Jan 21;18(1):51. doi: 10.1186/s12859-017-1481-7. BMC Bioinformatics. 2017. PMID: 28109252 Free PMC article.
-
CENTROIDFOLD: a web server for RNA secondary structure prediction.Nucleic Acids Res. 2009 Jul;37(Web Server issue):W277-80. doi: 10.1093/nar/gkp367. Epub 2009 May 12. Nucleic Acids Res. 2009. PMID: 19435882 Free PMC article.
-
RADAR: a web server for RNA data analysis and research.Nucleic Acids Res. 2007 Jul;35(Web Server issue):W300-4. doi: 10.1093/nar/gkm253. Epub 2007 May 21. Nucleic Acids Res. 2007. PMID: 17517784 Free PMC article.
-
ModeRNA server: an online tool for modeling RNA 3D structures.Bioinformatics. 2011 Sep 1;27(17):2441-2. doi: 10.1093/bioinformatics/btr400. Epub 2011 Jul 4. Bioinformatics. 2011. PMID: 21727140
-
Energy-based RNA consensus secondary structure prediction in multiple sequence alignments.Methods Mol Biol. 2014;1097:125-41. doi: 10.1007/978-1-62703-709-9_7. Methods Mol Biol. 2014. PMID: 24639158 Review.
Cited by
-
Biphasic patterns of diversification and the emergence of modules.Front Genet. 2012 Aug 7;3:147. doi: 10.3389/fgene.2012.00147. eCollection 2012. Front Genet. 2012. PMID: 22891076 Free PMC article.
-
Computational Characterization of Exogenous MicroRNAs that Can Be Transferred into Human Circulation.PLoS One. 2015 Nov 3;10(11):e0140587. doi: 10.1371/journal.pone.0140587. eCollection 2015. PLoS One. 2015. Retraction in: PLoS One. 2022 May 9;17(5):e0268437. doi: 10.1371/journal.pone.0268437. PMID: 26528912 Free PMC article. Retracted.
-
Menzerath-Altmann's Law of Syntax in RNA Accretion History.Life (Basel). 2021 May 27;11(6):489. doi: 10.3390/life11060489. Life (Basel). 2021. PMID: 34071925 Free PMC article.
-
Dietary MicroRNA Database (DMD): An Archive Database and Analytic Tool for Food-Borne microRNAs.PLoS One. 2015 Jun 1;10(6):e0128089. doi: 10.1371/journal.pone.0128089. eCollection 2015. PLoS One. 2015. PMID: 26030752 Free PMC article.
-
Phylogenetic study of nine species of freshwater monogeneans using secondary structure and motif prediction from India.Bioinformation. 2012;8(18):862-9. doi: 10.6026/97320630008862. Epub 2012 Sep 21. Bioinformation. 2012. PMID: 23144541 Free PMC article.
References
-
- Hermann T, Patel DJ. Stitching together RNA tertiary structures. J. Mol. Biol. 1999;294:829–849. - PubMed
-
- Billoud B, Guerrucci MA, Masselot M, Deutsch JS. Cirripede phylogeny using a novel approach: molecular morphometrics. Mol. Biol. Evol. 2000;17:1435–1445. - PubMed
-
- Collins LJ, Moulton V, Penny D. Use of RNA secondary structure for studying the evolution of RNase P and RNase MRP. J. Mol. Evol. 2000;51:194–2004. - PubMed
-
- Caetano-Anollés G. Novel strategies to study the role of mutation and nucleic acid structure in evolution. Plant Cell Tissue Organ Cult. 2001;67:115–132.

