The temperate marine phage PhiHAP-1 of Halomonas aquamarina possesses a linear plasmid-like prophage genome
- PMID: 18448537
- PMCID: PMC2447096
- DOI: 10.1128/JVI.00140-08
The temperate marine phage PhiHAP-1 of Halomonas aquamarina possesses a linear plasmid-like prophage genome
Abstract
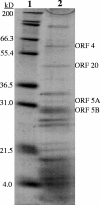
A myovirus-like temperate phage, PhiHAP-1, was induced with mitomycin C from a Halomonas aquamarina strain isolated from surface waters in the Gulf of Mexico. The induced cultures produced significantly more virus-like particles (VLPs) (3.73 x 10(10) VLP ml(-1)) than control cultures (3.83 x 10(7) VLP ml(-1)) when observed with epifluorescence microscopy. The induced phage was sequenced by using linker-amplified shotgun libraries and contained a genome 39,245 nucleotides in length with a G+C content of 59%. The PhiHAP-1 genome contained 46 putative open reading frames (ORFs), with 76% sharing significant similarity (E value of <10(-3)) at the protein level with other sequences in GenBank. Putative functional gene assignments included small and large terminase subunits, capsid and tail genes, an N6-DNA adenine methyltransferase, and lysogeny-related genes. Although no integrase was found, the PhiHAP-1 genome contained ORFs similar to protelomerase and parA genes found in linear plasmid-like phages with telomeric ends. Southern probing and PCR analysis of host genomic, plasmid, and PhiHAP-1 DNA indicated a lack of integration of the prophage with the host chromosome and a difference in genome arrangement between the prophage and virion forms. The linear plasmid prophage form of PhiHAP-1 begins with the protelomerase gene, presumably due to the activity of the protelomerase, while the induced phage particle has a circularly permuted genome that begins with the terminase genes. The PhiHAP-1 genome shares synteny and gene similarity with coliphage N15 and vibriophages VP882 and VHML, suggesting an evolutionary heritage from an N15-like linear plasmid prophage ancestor.
Figures







References
-
- Ackermann, H. W., and M. S. DuBow. 1987. Viruses of prokaryotes. CRC Press, Boca Raton, FL.
-
- Reference deleted.
-
- Bazinet, C., and J. King. 1985. The DNA translocating vertex of dsDNA bacteriophage. Annu. Rev. Microbiol. 39109-129. - PubMed
-
- Bergh, O., K. Y. Borsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in aquatic environments. Nature 340467-468. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

