Small interfering RNAs that deplete the cellular translation factor eIF4H impede mRNA degradation by the virion host shutoff protein of herpes simplex virus
- PMID: 18448541
- PMCID: PMC2447072
- DOI: 10.1128/JVI.00137-08
Small interfering RNAs that deplete the cellular translation factor eIF4H impede mRNA degradation by the virion host shutoff protein of herpes simplex virus
Abstract
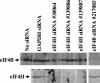
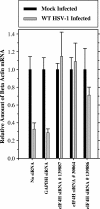
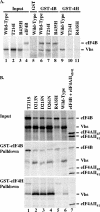
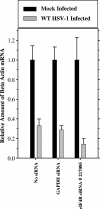
The herpes simplex virus (HSV) virion host shutoff (Vhs) protein is an endoribonuclease that accelerates decay of many host and viral mRNAs. Purified Vhs does not distinguish mRNAs from nonmessenger RNAs and cuts target RNAs at many sites, yet within infected cells it is targeted to mRNAs and cleaves those mRNAs at preferred sites including, for some, regions of translation initiation. This targeting may result in part from Vhs binding to the translation initiation factor eIF4H; in particular, several mutations in Vhs that abrogate its binding to eIF4H also abolish its mRNA-degradative activity, even though the mutant proteins retain endonuclease activity. To further investigate the role of eIF4H in Vhs activity, HeLa cells were depleted of eIF4H or other proteins by transfection with small interfering RNAs (siRNAs) 48 h prior to infection or mock infection in the presence of actinomycin D. Cellular mRNA levels were then assayed 5 h after infection. In cells transfected with an siRNA for the housekeeping enzyme glyceraldehyde-3-phosphate dehydrogenase, wild-type HSV infection reduced beta-actin mRNA levels to between 20 and 30% of those in mock-infected cells, indicative of a normal Vhs activity. In contrast, in cells transfected with any of three eIF4H siRNAs, beta-actin mRNA levels were indistinguishable in infected and mock-infected cells, suggesting that eIF4H depletion impeded Vhs-mediated degradation. Depletion of the related factor eIF4B did not affect Vhs activity. The data suggest that eIF4H binding is required for Vhs-induced degradation of many mRNAs, perhaps by targeting Vhs to mRNAs and to preferred sites within mRNAs.
Figures

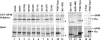

References
-
- Barzilai, A., I. Zivony-Elbom, R. Sarid, E. Noah, and N. Frenkel. 2006. The herpes simplex virus type 1 vhs-UL41 gene secures viral replication by temporarily evading apoptotic cellular response to infection: Vhs-UL41 activity might require interactions with elements of cellular mRNA degradation machinery. J. Virol. 80505-513. - PMC - PubMed
-
- Becker, Y., E. Tavor, Y. Asher, C. Berkowiltz, and M. Moyal. 1993. Effect of herpes simplex virus type-1 UL41 gene on the stability of mRNA from the cellular genes: beta-actin, fibronectin, glucose transporter-1, and docking protein, and on virus intraperitoneal pathogenicity of newborn mice. Virus Genes 7133-143. - PubMed
-
- Bevington, P. R. 1969. Data reduction and error analysis for the physical sciences. McGraw-Hill Book Company, New York, NY.
-
- Bookout, A. L., C. L. Cummins, D. J. Mangelsdorf, J. M. Pesola, and M. F. Kramer. 2006. High-throughput real-time quantitative reverse transcription PCR, p. 15.8.1-15.8.28. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

