Memory T cells specific for novel human papillomavirus type 16 (HPV16) E6 epitopes in women whose HPV16 infection has become undetectable
- PMID: 18448624
- PMCID: PMC2446617
- DOI: 10.1128/CVI.00404-07
Memory T cells specific for novel human papillomavirus type 16 (HPV16) E6 epitopes in women whose HPV16 infection has become undetectable
Abstract
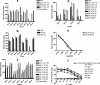
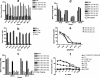
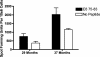
Human papillomavirus (HPV)-specific T-cell response to the HPV type 16 (HPV16) E6 protein has been shown to be associated with successful viral clearance. The patterns of CD8 T-cell epitopes within HPV16 E6 protein were previously studied in two women with HPV16 clearance. The goal of this study was to characterize these epitopes in terms of their minimal and optimal amino acid sequences and the human leukocyte antigen (HLA) restriction molecules. The presence of the epitope-specific memory T cells after viral clearance was also examined. In subject A, the dominant epitope was characterized to be E6 75-83 (KFYSKISEY), restricted by the HLA-B62 molecule, while that of subject B was E6 133-142 (HNIRGRWTGR), restricted by the HLA-A6801 molecule. Homologous epitopes were identified in five other high-risk HPV types for both of these epitopes, but they were not recognized by respective T-cell clone cells. An enzyme-linked immunospot assay or tetramer analysis was performed on peripheral blood mononuclear cells from blood samples collected after viral clearance but prior to isolation of the T-cell clones. The presence of epitope-specific memory T cells was demonstrated. These data suggest that HPV-specific memory T cells were generated in vivo and that they may remain in circulation many months, if not years, after viral clearance. Our findings broaden the spectrum of the CD8 T-cell epitopes of the HPV16 E6 protein. The characterization of novel T-cell epitopes and long-lasting epitope-specific memory T cells may be useful for the development of a potential epitope-based vaccine.
Figures
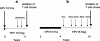
Similar articles
-
Human papillomavirus 16-specific T cell responses in classic HPV-related vulvar intra-epithelial neoplasia. Determination of strongly immunogenic regions from E6 and E7 proteins.Clin Exp Immunol. 2010 Jan;159(1):45-56. doi: 10.1111/j.1365-2249.2009.04006.x. Epub 2009 Jul 30. Clin Exp Immunol. 2010. PMID: 19843089 Free PMC article.
-
HLA class I binding promiscuity of the CD8 T-cell epitopes of human papillomavirus type 16 E6 protein.J Virol. 2007 Feb;81(3):1412-23. doi: 10.1128/JVI.01768-06. Epub 2006 Nov 15. J Virol. 2007. PMID: 17108051 Free PMC article.
-
Identification of HLA-DR1- and HLA-DR15-restricted human papillomavirus type 16 (HPV16) and HPV18 E6 epitopes recognized by CD4+ T cells from healthy young women.J Gen Virol. 2007 May;88(Pt 5):1470-1478. doi: 10.1099/vir.0.82558-0. J Gen Virol. 2007. PMID: 17412975
-
Identification of an HLA-A24-restricted cytotoxic T lymphocyte epitope from human papillomavirus type-16 E6: the combined effects of bortezomib and interferon-gamma on the presentation of a cryptic epitope.Int J Cancer. 2007 Feb 1;120(3):594-604. doi: 10.1002/ijc.22312. Int J Cancer. 2007. PMID: 17096336
-
Development of a Spontaneous HPV16 E6/E7-Expressing Head and Neck Squamous Cell Carcinoma in HLA-A2 Transgenic Mice.mBio. 2022 Feb 22;13(1):e0325221. doi: 10.1128/mbio.03252-21. Epub 2022 Jan 4. mBio. 2022. PMID: 35089069 Free PMC article.
Cited by
-
A novel prostate cancer immunotherapy using prostate-specific antigen peptides and Candida skin test reagent as an adjuvant.SAGE Open Med. 2018 Sep 17;6:2050312118800202. doi: 10.1177/2050312118800202. eCollection 2018. SAGE Open Med. 2018. PMID: 30245818 Free PMC article.
-
Biomarkers of oxidant load and type-specific clearance of prevalent oncogenic human papillomavirus infection: markers of immune response?Int J Cancer. 2012 Jul 1;131(1):219-28. doi: 10.1002/ijc.26363. Epub 2011 Nov 28. Int J Cancer. 2012. PMID: 21858808 Free PMC article.
-
CD4+ T cells against human papillomavirus-18 E7 in patients with high-grade cervical lesions associate with the absence of the virus in the cervix.Immunology. 2010 Sep;131(1):89-98. doi: 10.1111/j.1365-2567.2010.03277.x. Epub 2010 Jun 2. Immunology. 2010. PMID: 20545782 Free PMC article.
-
A Human Papillomavirus Type 16 E6 52-62 CD4 T-Cell Epitope Restricted by the HLA-DR11 Molecule Described in an Epitope Hotspot.MOJ Immunol. 2014;1(3):00018. doi: 10.15406/moji.2014.01.00018. MOJ Immunol. 2014. PMID: 25685851 Free PMC article.
-
Human papillomavirus 16-specific T cell responses in classic HPV-related vulvar intra-epithelial neoplasia. Determination of strongly immunogenic regions from E6 and E7 proteins.Clin Exp Immunol. 2010 Jan;159(1):45-56. doi: 10.1111/j.1365-2249.2009.04006.x. Epub 2009 Jul 30. Clin Exp Immunol. 2010. PMID: 19843089 Free PMC article.
References
-
- Alexander, M., M. L. Salgaller, E. Celis, A. Sette, W. A. Barnes, S. A. Rosenberg, and M. A. Steller. 1996. Generation of tumor-specific cytolytic T lymphocytes from peripheral blood of cervical cancer patients by in vitro stimulation with a synthetic human papillomavirus type 16 E7 epitope. Am. J. Obstet. Gynecol. 175:1586-1593. - PubMed
-
- Bontkes, H. J., T. D. de Gruijl, J. M. Walboomers, A. J. van den Muysenberg, A. W. Gunther, R. J. Scheper, C. J. Meijer, and J. A. Kummer. 1997. Assessment of cytotoxic T-lymphocyte phenotype using the specific markers granzyme B and TIA-1 in cervical neoplastic lesions. Br. J. Cancer 76:1353-1360. - PMC - PubMed
-
- Bourgault Villada, I., N. Beneton, C. Bony, F. Connan, J. Monsonego, A. Bianchi, P. Saiag, J. P. Levy, J. G. Guillet, and J. Choppin. 2000. Identification in humans of HPV-16 E6 and E7 protein epitopes recognized by cytolytic T lymphocytes in association with HLA-B18 and determination of the HLA-B18-specific binding motif. Eur. J. Immunol. 30:2281-2289. - PubMed
-
- Crook, T., J. A. Tidy, and K. H. Vousden. 1991. Degradation of p53 can be targeted by HPV E6 sequences distinct from those required for p53 binding and trans-activation. Cell 67:547-556. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

