Method To Detect Only Live Bacteria during PCR Amplification
- PMID: 18448692
- PMCID: PMC2446937
- DOI: 10.1128/JCM.02171-07
Method To Detect Only Live Bacteria during PCR Amplification
Abstract
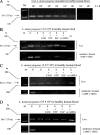
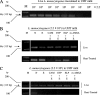
Ethidium monoazide (EMA) is a DNA cross-linking agent and eukaryotic topoisomerase II poison. We previously reported that the treatment of EMA with visible light irradiation (EMA + Light) directly cleaved chromosomal DNA of Escherichia coli (T. Soejima, K. Iida, T. Qin, H. Taniai, M. Seki, A. Takade, and S. Yoshida, Microbiol. Immunol. 51:763-775, 2007). Herein, we report that EMA + Light randomly cleaved chromosomal DNA of heat-treated, but not live, Listeria monocytogenes cells within 10 min of treatment. When PCR amplified DNA that was 894 bp in size, PCR final products from 10(8) heat-treated L. monocytogenes were completely suppressed by EMA + Light. When target DNA was short (113 bp), like the hly gene of L. monocytogenes, DNA amplification was not completely suppressed by EMA + Light only. Thus, we used DNA gyrase/topoisomerase IV and mammalian topoisomerase poisons (here abbreviated as T-poisons) together with EMA + Light. T-poisons could penetrate heat-treated, but not live, L. monocytogenes cells within 30 min to cleave chromosomal DNA by poisoning activity. The PCR product of the hly gene from 10(8) heat-treated L. monocytogenes cells was inhibited by a combination of EMA + Light and T-poisons (EMA + Light + T-poisons), but those from live bacteria were not suppressed. As a model for clinical application to bacteremia, we tried to discriminate live and antibiotic-treated L. monocytogenes cells present in human blood. EMA + Light + T-poisons completely suppressed the PCR product from 10(3) to 10(7) antibiotic-treated L. monocytogenes cells but could detect 10(2) live bacteria. Considering the prevention and control of food poisoning, this method was applied to discriminate live and heat-treated L. monocytogenes cells spiked into pasteurized milk. EMA + Light + T-poisons inhibited the PCR product from 10(3) to 10(7) heat-treated cells but could detect 10(1) live L. monocytogenes cells. Our method is useful in clinical as well as food hygiene tests.
Figures





Similar articles
-
Photoactivated ethidium monoazide directly cleaves bacterial DNA and is applied to PCR for discrimination of live and dead bacteria.Microbiol Immunol. 2007;51(8):763-75. doi: 10.1111/j.1348-0421.2007.tb03966.x. Microbiol Immunol. 2007. PMID: 17704639
-
Insufficient differentiation of live and dead Campylobacter jejuni and Listeria monocytogenes cells by ethidium monoazide (EMA) compromises EMA/real-time PCR.Res Microbiol. 2007 Jun;158(5):405-12. doi: 10.1016/j.resmic.2007.02.008. Epub 2007 Mar 12. Res Microbiol. 2007. PMID: 17449228
-
Polymerase chain reaction amplification length-dependent ethidium monoazide suppression power for heat-killed cells of Enterobacteriaceae.Anal Biochem. 2011 Nov 1;418(1):37-43. doi: 10.1016/j.ab.2011.06.027. Epub 2011 Jun 29. Anal Biochem. 2011. PMID: 21771573
-
The exclusive use of flow cytometry to evaluate the antibiotic-susceptibility.Biochim Biophys Acta. 2012 Dec;1820(12):1980-6. doi: 10.1016/j.bbagen.2012.09.003. Epub 2012 Sep 12. Biochim Biophys Acta. 2012. PMID: 22982588
-
An advanced PCR method for the specific detection of viable total coliform bacteria in pasteurized milk.Appl Microbiol Biotechnol. 2012 Jul;95(2):485-97. doi: 10.1007/s00253-012-4086-0. Epub 2012 May 30. Appl Microbiol Biotechnol. 2012. PMID: 22644523
Cited by
-
Method to quantify live and dead cells in multi-species oral biofilm by real-time PCR with propidium monoazide.AMB Express. 2013 Jan 4;3(1):1. doi: 10.1186/2191-0855-3-1. AMB Express. 2013. PMID: 23289803 Free PMC article.
-
Viable and Total Bacterial Populations Undergo Equipment- and Time-Dependent Shifts during Milk Processing.Appl Environ Microbiol. 2019 Jun 17;85(13):e00270-19. doi: 10.1128/AEM.00270-19. Print 2019 Jul 1. Appl Environ Microbiol. 2019. PMID: 31028031 Free PMC article.
-
Validation of the efficacy of air purifiers using molecular techniques.PLoS One. 2023 Jan 9;18(1):e0280243. doi: 10.1371/journal.pone.0280243. eCollection 2023. PLoS One. 2023. PMID: 36622844 Free PMC article.
-
Viability PCR, a culture-independent method for rapid and selective quantification of viable Legionella pneumophila cells in environmental water samples.Appl Environ Microbiol. 2009 Jun;75(11):3502-12. doi: 10.1128/AEM.02878-08. Epub 2009 Apr 10. Appl Environ Microbiol. 2009. PMID: 19363080 Free PMC article.
-
Development of a sensitive and specific qPCR assay in conjunction with propidium monoazide for enhanced detection of live Salmonella spp. in food.BMC Microbiol. 2013 Dec 1;13:273. doi: 10.1186/1471-2180-13-273. BMC Microbiol. 2013. PMID: 24289661 Free PMC article.
References
-
- Adesiyun, A. A. 1994. Bacteriological quality and associated public health risk of pre-processed bovine milk in Trinidad. Int. J. Food Microbiol. 21253-261. - PubMed
-
- Almeida, G., A. Figueiredo, M. Rôla, R. M. Barros, P. Gibbs, T. Hogg, and P. Teixeira. 2007. Microbiolgical characterization of randomly selected Portuguese raw milk cheeses with reference to food safety. J. Food Prot. 701710-1716. - PubMed
-
- Burden, D. A., P. S. Kingma, S. J. Froelich-Ammon, M.-A. Bjornsti, M. W. Patchan, R. B. Thompson, and N. Osheroff. 1996. Topoisomerase II·etoposide interactions direct the formation of drug-induced enzyme-DNA cleavage complexes. J. Biol. Chem. 27129238-29244. - PubMed
-
- Dall'Acqua, F., D. Vedaldi, S. Caffieri, A. Guiotto, P. Rodighiero, F. Baccichetti, F. Carlassare, and F. Bordin. 1981. New monofunctional reagents for DNA as possible agents for the photochemotherapy of psoriasis: derivatives of 4,5′-dimethylangelicin. J. Med. Chem. 24178-184. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

