Efficient entry inhibition of human and nonhuman primate immunodeficiency virus by cell surface-expressed gp41-derived peptides
- PMID: 18449216
- PMCID: PMC2862551
- DOI: 10.1038/gt.2008.73
Efficient entry inhibition of human and nonhuman primate immunodeficiency virus by cell surface-expressed gp41-derived peptides
Abstract
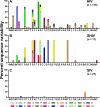
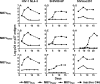
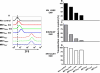
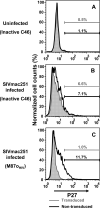
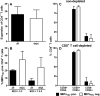
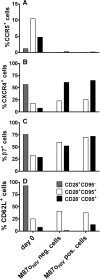
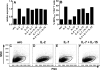
Membrane-anchored C-peptides (for example, maC46) derived from human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein gp41 effectively inhibit HIV-1 entry in cell lines and primary human CD4+ cells in vitro. Here we evaluated this gene therapy approach in animal models of AIDS. We adapted the HIV gp41-derived maC46 vector construct for use in rhesus monkeys. Simian immunodeficiency virus (SIV and SHIV) sequence-adapted maC46 peptides, and the original HIV-1-derived maC46 expressed on the surface of established cell lines blocked entry of HIV-1, SIVmac251 and SHIV89.6P. Furthermore, primary rhesus monkey CD4+ T cells expressing HIV sequence-based maC46 peptides were also protected from SIV entry. Depletion of CD8+ T cells from PBMCs enhanced the yield of maC46-transduced CD4+ T cells. Supplementation with interleukin-2 (IL-2) increased transduction efficiency, whereas IL-7 and/or IL-15 provided no additional benefit. Phenotypic analysis showed that maC46-transduced and expanded cells were predominantly central memory CD4+ T cells that expressed low levels of CCR5 and slightly elevated levels of CD62L, beta7-integrin and CXCR4. These findings show that maC46-based cell surface-expressed peptides can efficiently inhibit primate immunodeficiency virus infection, and therefore serve as the basis for evaluation of this gene therapy approach in an animal model for AIDS.
Figures
References
-
- Puls RL, Emery S. Therapeutic vaccination against HIV: current progress and future possibilities. Clin Sci. 2006;110:59–71. - PubMed
-
- von Laer D, Hasselmann S, Hasselmann K. Gene therapy for HIV infection: what does it need to make it work? J Gene Med. 2006;8:658–667. - PubMed
-
- Strayer DS, Akkina R, Bunnell BA, Dropulic B, Planelles V, Pomerantz RJ, et al. Current status of gene therapy strategies to treat HIV/AIDS. Mol Ther. 2005;11:823–842. - PubMed
-
- von Laer D, Hasselmann S, Hasselmann K. Impact of gene-modified T cells on HIV infection dynamics. J Theor Biol. 2006;238:60–77. - PubMed
-
- Gallo SA, Finnegan CM, Viard M, Raviv Y, Dimitrov A, Rawat SS, et al. The HIV Env-mediated fusion reaction. Biochim Biophys Acta. 2003;1614:36–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

