Protein phosphatase-1 inhibitor-3 is an in vivo target of caspase-3 and participates in the apoptotic response
- PMID: 18450750
- PMCID: PMC2440612
- DOI: 10.1074/jbc.M709735200
Protein phosphatase-1 inhibitor-3 is an in vivo target of caspase-3 and participates in the apoptotic response
Abstract
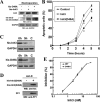
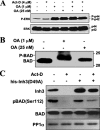
Inh3 (inhibitor-3) is a potent inhibitor of protein phosphatase-1 that selectively associates with PP1gamma1 and PP1alpha but not the PP1beta isoform. We demonstrate that Inh3 is a novel substrate for caspase-3 and is degraded in vivo during apoptosis induced by actinomycin D. Inh3 was not degraded in apoptotic MCF-7 cells, which lack caspase-3. These experiments establish that Inh3 is a novel physiological substrate of caspase-3. Electroporation of the caspase-3-resistant Inh3-D49A mutant into HL-60 cells resulted in a significant attenuation of apoptosis induced by actinomycin D. These results show that Inh3 degradation contributes to the apoptotic process. Immunofluorescence based examination of the subcellular localizations of Inh3 and PP1gamma1 revealed a major relocalization of the cellular pool of PP1gamma1 from the nucleolus to the nucleus and then to the cytoplasm during actinomycin D-induced apoptosis. A similar redistribution of PP1alpha from the nucleus to the cytoplasm occurred. These results are consistent with an unexpected discovery that significant fractions of the cellular pools of PP1gamma1 and PP1alpha are associated with Inh3 in HL-60 cells. Thus, Inh3 is a major factor in the cellular economy of PP1gamma1 and PP1alpha subunits. The unscheduled relocalization of this large a pool of PP1 subunits and their release from a potent inhibitor could deregulate a diverse range of essential cellular processes and signaling pathways. We discuss the significance of these findings in relation to working hypotheses whereby Inh3 destruction could contribute to the apoptotic process.
Figures






References
-
- Ceulemans, H., and Bollen, M. (2004) Physiol. Rev. 84 1-39 - PubMed
-
- Cohen, P. (1989) Annu. Rev. Biochem. 58 453-508 - PubMed
-
- Lee, E. Y., Zhang, L., Zhao, S., Wei, Q., Zhang, J., Qi, Z. Q., and Belmonte, E. R. (1999) Front. Biosci. 4 D270-D285 - PubMed
-
- Shima, H., Hatano, Y., Chun, Y. S., Sugimura, T., Zhang, Z., Lee, E. Y., and Nagao, M. (1993) Biochem. Biophys. Res. Commun. 192 1289-1296 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

