Rational design of human DNA ligase inhibitors that target cellular DNA replication and repair
- PMID: 18451142
- PMCID: PMC2734474
- DOI: 10.1158/0008-5472.CAN-07-6636
Rational design of human DNA ligase inhibitors that target cellular DNA replication and repair
Abstract
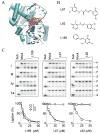
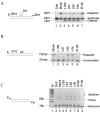
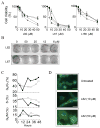
Based on the crystal structure of human DNA ligase I complexed with nicked DNA, computer-aided drug design was used to identify compounds in a database of 1.5 million commercially available low molecular weight chemicals that were predicted to bind to a DNA-binding pocket within the DNA-binding domain of DNA ligase I, thereby inhibiting DNA joining. Ten of 192 candidates specifically inhibited purified human DNA ligase I. Notably, a subset of these compounds was also active against the other human DNA ligases. Three compounds that differed in their specificity for the three human DNA ligases were analyzed further. L82 inhibited DNA ligase I, L67 inhibited DNA ligases I and III, and L189 inhibited DNA ligases I, III, and IV in DNA joining assays with purified proteins and in cell extract assays of DNA replication, base excision repair, and nonhomologous end-joining. L67 and L189 are simple competitive inhibitors with respect to nicked DNA, whereas L82 is an uncompetitive inhibitor that stabilized complex formation between DNA ligase I and nicked DNA. In cell culture assays, L82 was cytostatic whereas L67 and L189 were cytotoxic. Concordant with their ability to inhibit DNA repair in vitro, subtoxic concentrations of L67 and L189 significantly increased the cytotoxicity of DNA-damaging agents. Interestingly, the ligase inhibitors specifically sensitized cancer cells to DNA damage. Thus, these novel human DNA ligase inhibitors will not only provide insights into the cellular function of these enzymes but also serve as lead compounds for the development of anticancer agents.
Figures
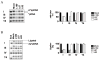

Similar articles
-
Structure-activity relationships among DNA ligase inhibitors: Characterization of a selective uncompetitive DNA ligase I inhibitor.DNA Repair (Amst). 2017 Dec;60:29-39. doi: 10.1016/j.dnarep.2017.10.002. Epub 2017 Oct 10. DNA Repair (Amst). 2017. PMID: 29078112 Free PMC article.
-
Identification and validation of human DNA ligase inhibitors using computer-aided drug design.J Med Chem. 2008 Aug 14;51(15):4553-62. doi: 10.1021/jm8001668. Epub 2008 Jul 17. J Med Chem. 2008. PMID: 18630893 Free PMC article.
-
Impact of DNA ligase inhibition on the nick sealing of polβ nucleotide insertion products at the downstream steps of base excision repair pathway.Mutagenesis. 2024 Nov 2;39(6):263-279. doi: 10.1093/mutage/geae013. Mutagenesis. 2024. PMID: 38736258 Free PMC article.
-
Mammalian DNA ligases; roles in maintaining genome integrity.J Mol Biol. 2024 Jan 1;436(1):168276. doi: 10.1016/j.jmb.2023.168276. Epub 2023 Sep 13. J Mol Biol. 2024. PMID: 37714297 Free PMC article. Review.
-
DNA ligases in the repair and replication of DNA.Mutat Res. 2000 Aug 30;460(3-4):301-18. doi: 10.1016/s0921-8777(00)00033-1. Mutat Res. 2000. PMID: 10946235 Review.
Cited by
-
Unchanged PCNA and DNMT1 dynamics during replication in DNA ligase I-deficient cells but abnormal chromatin levels of non-replicative histone H1.Sci Rep. 2023 Mar 16;13(1):4363. doi: 10.1038/s41598-023-31367-4. Sci Rep. 2023. PMID: 36928068 Free PMC article.
-
A high-throughput small molecule screen identifies farrerol as a potentiator of CRISPR/Cas9-mediated genome editing.Elife. 2020 Jul 9;9:e56008. doi: 10.7554/eLife.56008. Elife. 2020. PMID: 32644042 Free PMC article.
-
Human DNA Ligase I Interacts with and Is Targeted for Degradation by the DCAF7 Specificity Factor of the Cul4-DDB1 Ubiquitin Ligase Complex.J Biol Chem. 2016 Oct 14;291(42):21893-21902. doi: 10.1074/jbc.M116.746198. Epub 2016 Aug 29. J Biol Chem. 2016. PMID: 27573245 Free PMC article.
-
Translational research in radiation-induced DNA damage signaling and repair.Transl Cancer Res. 2017 Jul;6(Suppl 5):S875-S891. doi: 10.21037/tcr.2017.06.02. Transl Cancer Res. 2017. PMID: 30574452 Free PMC article.
-
DNA damage repair: historical perspectives, mechanistic pathways and clinical translation for targeted cancer therapy.Signal Transduct Target Ther. 2021 Jul 9;6(1):254. doi: 10.1038/s41392-021-00648-7. Signal Transduct Target Ther. 2021. PMID: 34238917 Free PMC article. Review.
References
-
- Madhusudan S, Hickson ID. DNA repair inhibition: a selective tumour targeting strategy. Trends Mol Med. 2005;11:503–11. - PubMed
-
- Tomkinson AE, Vijayakumar S, Pascal JM, Ellenberger T. DNA ligases: structure, reaction mechanism, and function. Chem Rev. 2006;106:687–99. - PubMed
-
- Sun D, Urrabaz R. Development of non-electrophoretic assay method for DNA ligases and its application to screening of chemical inhibitors of DNA ligase I. J Biochem Biophys Methods. 2004;59:49–59. - PubMed
-
- Pascal JM, O’Brien PJ, Tomkinson AE, Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432:473–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM052504/GM/NIGMS NIH HHS/United States
- CA92584/CA/NCI NIH HHS/United States
- R01 ES012512/ES/NIEHS NIH HHS/United States
- ES12512/ES/NIEHS NIH HHS/United States
- P01 CA092584/CA/NCI NIH HHS/United States
- R01 GM057479/GM/NIGMS NIH HHS/United States
- R01 GM047251/GM/NIGMS NIH HHS/United States
- GM52504/GM/NIGMS NIH HHS/United States
- R01 CA102428/CA/NCI NIH HHS/United States
- GM57479/GM/NIGMS NIH HHS/United States
- CA102428/CA/NCI NIH HHS/United States
- R56 CA102428/CA/NCI NIH HHS/United States
- GM47251/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

