Sequential transcription factor targeting for diffuse large B-cell lymphomas
- PMID: 18451163
- PMCID: PMC2748725
- DOI: 10.1158/0008-5472.CAN-07-5817
Sequential transcription factor targeting for diffuse large B-cell lymphomas
Abstract
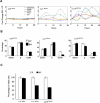
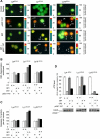
Transcription factors play a central role in malignant transformation by activating or repressing waves of downstream target genes. Therapeutic targeting of transcription factors can reprogram cancer cells to lose their advantages in growth and survival. The BCL6 transcriptional repressor plays a central role in the pathogenesis of diffuse large B-cell lymphomas (DLBCL) and controls downstream checkpoints, including the p53 tumor suppressor gene. We report that a specific inhibitor of BCL6 called BPI can trigger a p53 response in DLBCL cells. This was partially due to induction of p53 activity and partially due to relief of direct repression by BCL6 of p53 target genes. BPI could thus induce a p53-like response even in the presence of mutant p53. Moreover, sequential BCL6 peptide inhibitors followed by p53 peptide or small-molecule activators provided a more powerful antilymphoma effect than either treatment alone by maximally restoring p53 target gene expression. Therefore, tandem targeting of the overlapping BCL6 and p53 transcriptional programs can correct aberrant survival pathways in DLBCL and might provide an effective therapeutic approach to lymphoma therapy.
Figures




Similar articles
-
The Expanding Role of the BCL6 Oncoprotein as a Cancer Therapeutic Target.Clin Cancer Res. 2017 Feb 15;23(4):885-893. doi: 10.1158/1078-0432.CCR-16-2071. Epub 2016 Nov 23. Clin Cancer Res. 2017. PMID: 27881582 Free PMC article. Review.
-
BCL6 repression of EP300 in human diffuse large B cell lymphoma cells provides a basis for rational combinatorial therapy.J Clin Invest. 2010 Dec 1;120(12):4569-82. doi: 10.1172/JCI42869. Epub 2010 Nov 1. J Clin Invest. 2010. PMID: 21041953 Free PMC article.
-
A peptomimetic inhibitor of BCL6 with potent antilymphoma effects in vitro and in vivo.Blood. 2009 Apr 9;113(15):3397-405. doi: 10.1182/blood-2008-07-168773. Epub 2008 Oct 16. Blood. 2009. PMID: 18927431 Free PMC article.
-
Therapeutic targeting of the BCL6 oncogene for diffuse large B-cell lymphomas.Leuk Lymphoma. 2008 May;49(5):874-82. doi: 10.1080/10428190801895345. Leuk Lymphoma. 2008. PMID: 18452090 Free PMC article. Review.
-
Transcriptional signature with differential expression of BCL6 target genes accurately identifies BCL6-dependent diffuse large B cell lymphomas.Proc Natl Acad Sci U S A. 2007 Feb 27;104(9):3207-12. doi: 10.1073/pnas.0611399104. Epub 2007 Feb 20. Proc Natl Acad Sci U S A. 2007. PMID: 17360630 Free PMC article.
Cited by
-
iRGD-modified exosomes-delivered BCL6 siRNA inhibit the progression of diffuse large B-cell lymphoma.Front Oncol. 2022 Aug 2;12:822805. doi: 10.3389/fonc.2022.822805. eCollection 2022. Front Oncol. 2022. PMID: 35982974 Free PMC article.
-
The Expanding Role of the BCL6 Oncoprotein as a Cancer Therapeutic Target.Clin Cancer Res. 2017 Feb 15;23(4):885-893. doi: 10.1158/1078-0432.CCR-16-2071. Epub 2016 Nov 23. Clin Cancer Res. 2017. PMID: 27881582 Free PMC article. Review.
-
Ex vivo engineered immune organoids for controlled germinal center reactions.Biomaterials. 2015 Sep;63:24-34. doi: 10.1016/j.biomaterials.2015.06.002. Epub 2015 Jun 3. Biomaterials. 2015. PMID: 26072995 Free PMC article.
-
Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design.J Med Chem. 2018 Sep 13;61(17):7573-7588. doi: 10.1021/acs.jmedchem.8b00040. Epub 2018 Jul 17. J Med Chem. 2018. PMID: 29969259 Free PMC article.
-
Biomarkers and novel therapeutic approaches for diffuse large B-cell lymphoma in the era of precision medicine.Oncotarget. 2020 Nov 3;11(44):4045-4073. doi: 10.18632/oncotarget.27785. eCollection 2020 Nov 3. Oncotarget. 2020. PMID: 33216822 Free PMC article. Review.
References
-
- Pasqualucci L, Bereschenko O, Niu H, et al. Molecular pathogenesis of non-Hodgkin's lymphoma: the role of Bcl-6. Leuk Lymphoma. 2003;44(Suppl 3):S5–12. - PubMed
-
- Cattoretti G, Pasqualucci L, Ballon G, et al. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7(5):445–55. - PubMed
-
- Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432(7017):635–9. - PubMed
-
- Phan RT, Saito M, Basso K, Niu H, Dalla-Favera R. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat Immunol. 2005;6(10):1054–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

