PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows
- PMID: 18451766
- PMCID: PMC2373374
- DOI: 10.1038/embor.2008.56
PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows
Abstract
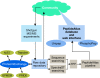
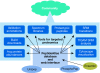
A crucial part of a successful systems biology experiment is an assay that provides reliable, quantitative measurements for each of the components in the system being studied. For proteomics to be a key part of such studies, it must deliver accurate quantification of all the components in the system for each tested perturbation without any gaps in the data. This will require a new approach to proteomics that is based on emerging targeted quantitative mass spectrometry techniques. The PeptideAtlas Project comprises a growing, publicly accessible database of peptides identified in many tandem mass spectrometry proteomics studies and software tools that allow the building of PeptideAtlas, as well as its use by the research community. Here, we describe the PeptideAtlas Project, its contents and components, and show how together they provide a unique platform to select and validate mass spectrometry targets, thereby allowing the next revolution in proteomics.
Figures



References
-
- Anderson L, Hunter CL (2006) Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics 5: 573–588 - PubMed
-
- Craig R, Beavis RC (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20: 1466–1467 - PubMed
-
- Craig R, Cortens JP, Beavis RC (2004) Open source system for analyzing, validating, and storing protein identification data. J Proteome Res 3: 1234–1242 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

