Localization and nucleotide specificity of Blastocystis succinyl-CoA synthetase
- PMID: 18452512
- PMCID: PMC2440562
- DOI: 10.1111/j.1365-2958.2008.06228.x
Localization and nucleotide specificity of Blastocystis succinyl-CoA synthetase
Abstract
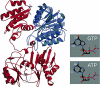
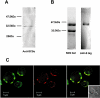
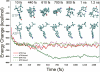
The anaerobic lifestyle of the intestinal parasite Blastocystis raises questions about the biochemistry and function of its mitochondria-like organelles. We have characterized the Blastocystis succinyl-CoA synthetase (SCS), a tricarboxylic acid cycle enzyme that conserves energy by substrate-level phosphorylation. We show that SCS localizes to the enigmatic Blastocystis organelles, indicating that these organelles might play a similar role in energy metabolism as classic mitochondria. Although analysis of residues inside the nucleotide-binding site suggests that Blastocystis SCS is GTP-specific, we demonstrate that it is ATP-specific. Homology modelling, followed by flexible docking and molecular dynamics simulations, indicates that while both ATP and GTP fit into the Blastocystis SCS active site, GTP is destabilized by electrostatic dipole interactions with Lys 42 and Lys 110, the side-chains of which lie outside the nucleotide-binding cavity. It has been proposed that residues in direct contact with the substrate determine nucleotide specificity in SCS. However, our results indicate that, in Blastocystis, an electrostatic gatekeeper controls which ligands can enter the binding site.
Figures
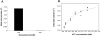
References
-
- Basu G, Sivanesan D, Kawabata T, Go N. Electrostatic potential of nucleotide-free protein is sufficient for discrimination between adenine and guanine-specific binding sites. J Mol Biol. 2004;342:1053–1066. - PubMed
-
- Bridger WA, Ramaley RF, Boyer PD, John ML. Succinyl-coenzyme A synthetase from Escherichia coli. Methods Enzymol. 1969;13:70–75.
-
- Bridger WA, Wolodko WT, Henning W, Upton C, Majumdar R, Williams SP. The subunits of succinyl-coenzyme A synthetase – function and assembly. Biochem Soc Symp. 1987;54:103–111. - PubMed
-
- Castillo-Davis CI, Mekhedov SL, Hartl DL, Koonin EV, Kondrashov FA. Selection for short introns in highly expressed genes. Nat Genet. 2002;31:415–418. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

